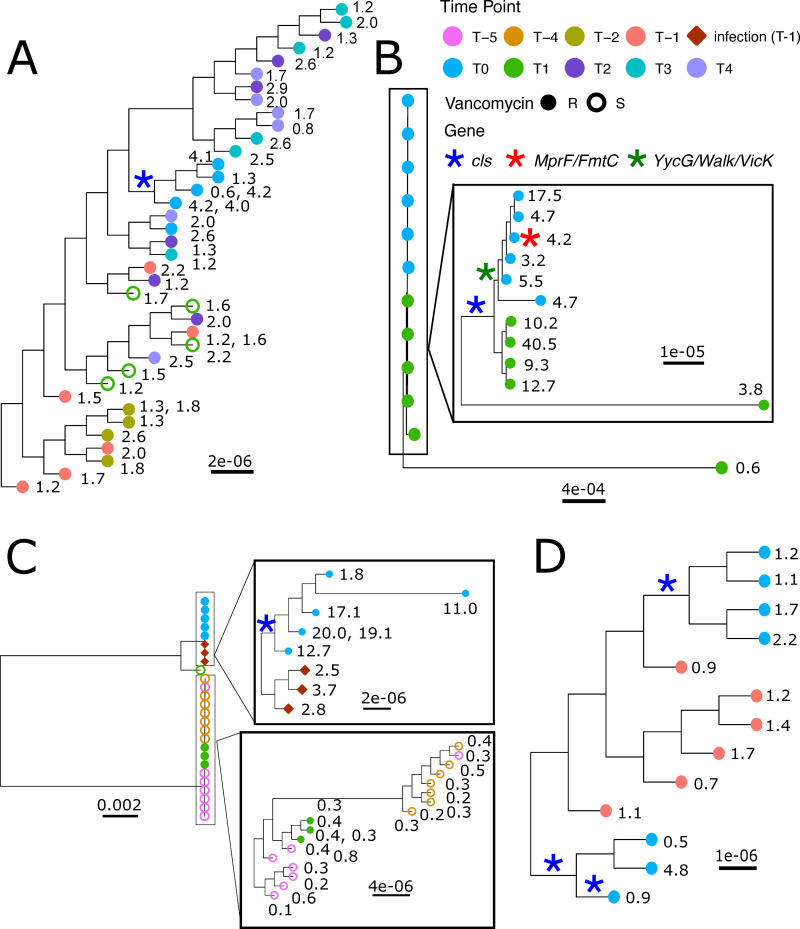
Fig 7. Individual phylogenies for E. faecium strains isolated from 4 patients.
A midpoint-rooted phylogeny was created based on the core genome alignment of all isolates using RAxML. Here, subsets of the full tree (Fig 6) are shown, including only the isolates from each patient (i.e., other tips are masked). Scale bars for branch length (mean number of substitutions per site) are included for each tree and insert. Rectangle inserts show detailed structure of indicated groups. Daptomycin MICCs are listed for each tip. For tips representing 2 isolates with identical core genomes, the daptomycin MICCs for both isolates are listed. Asterisks indicate mutations in genes known to be associated with variation in daptomycin susceptibility. For each patient, one assembled genome (two for the 2 clades in Patient 150) was used as a reference genome for variant calling (see Methods). (A) Patient 4 (B) Patient 86 (C) Patient 150 (D) Patient 87. Sequence data is available at NCBI GenBank under the study accession number PRJNA673360.