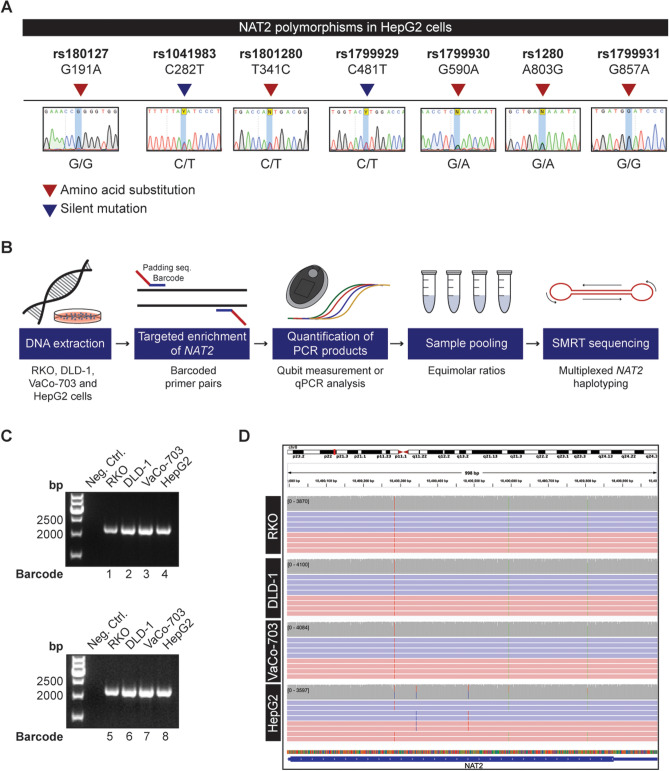
Figure 5.
NAT2 haplotype resolution by multiplex SMRT sequencing. (A) Sanger sequencing does not allow the resolution of NAT2 haplotypes in samples that are polymorphic in more than one position. The genotype of HepG2 cells is shown for each of the seven standard positions used for NAT2 allele calling. (B) Workflow for multiplexed SMRT sequencing. Genomic DNA was extracted from the cancer cell lines to be haplotyped. The NAT2 region is amplified in each sample by using primer pairs with a unique barcode and a padding sequence. The resulting PCR products were quantified by Qubit measurements or qPCR analysis and pooled in equimolar ratios for multiplexed SMRT sequencing. (C) Barcoded PCR products resulting from NAT2 amplification in RKO, DLD-1, VaCo-703 and HepG2 cell lines with two different sets of primers. Samples were run in a 1.5% agarose gel and water was used as a negative PCR control. Full-length gels are presented in Supplementary Fig. 4. (D) Visualization of the individual alleles present in RKO, DLD-1, VaCo-703 and HepG2 cell lines in Integrative Genomics Viewer. De-multiplexed SMRT sequencing reads from each cell line are aligned to the NAT2 reference sequence. Selection of reads in both positive (red) and negative (blue) direction supporting the NAT2*6A allele in RKO, DLD1, VaCo-703, HepG2 and NAT2*5B allele in HepG2 are shown. The gray stacks above each alignment represent the sequencing coverage. Vertical colored lines illustrate the positions of variation from the reference.