Figure 7.
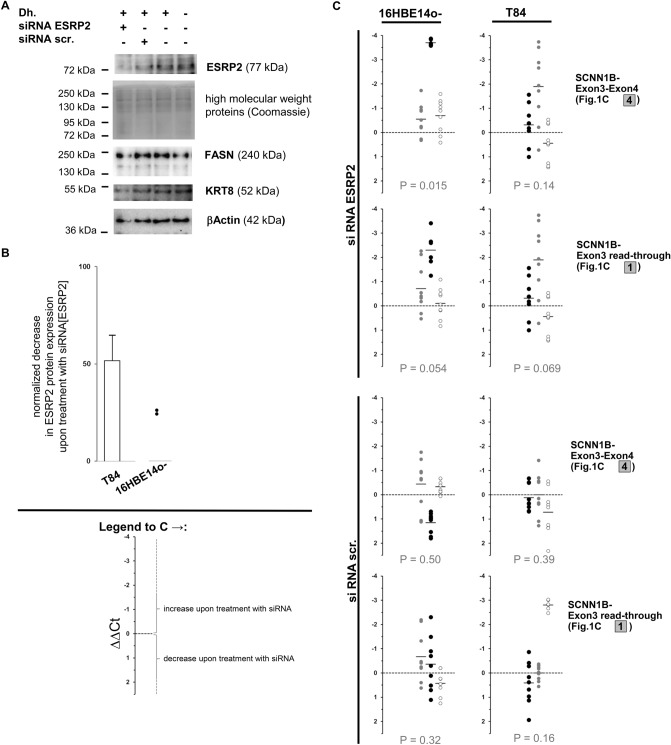
siRNA-mediated downregulation of ESRP2 affects the SCNN1B transcript spectrum in 16HBE14o-cells. siRNA directed against ESRP2 (siRNA ESRP2; mix of four siRNAs, on-target plus pool, GE Healthcare) or scrambled control (siRNA scr., GE Healthcare); was provided to T84 and 16HBE14o-cells. ESRP2 protein expression and SCNN1B mRNA expression levels were compared between cells supplied with the transfection agent Dharmafect 1 (Dh.) only versus cells that received Dharmafect and siRNA directed against ESRP2. Experiments in both cell lines were conducted from two (T84) or three (16HBE14o) different passages as biologically independent replicates whereby in each of these experiments, cell culture plates were treated in parallel for the following conditions: growth control (no Dharmafect transfection agent or siRNA; 8 plates), transfection agent control (with Dharmafect but without siRNA; 8 plates), treated with siRNA directed against ESRP2 (8 plates) and treated with scrambled siRNA (8 plates). Four out of eight plates were harvested at 24 h post-transfection and the remaining four plates were analyzed after 48 h post-transfection. Nuclear extracts were analyzed for ESRP2 protein expression from one plate in these sets of four (A,B). RNA was isolated from the remaining three plates (C). (A) Nuclear extracts derived from 16HBE14o-cells treated with 100 pmol siRNA directed against ESRP2 (siRNA ESRP2) or scrambled control (siRNA scr.), transfection control and growth control were analyzed for ESRP2 protein by western blot. The intensity of Coomassie-stained high molecular weight bands was used as a western blot loading control. Signals for fatty-acid synthase (FASN), keratin 8 (KRT8) and βActin are provided as control detections. (B) Reduction of protein expression levels of ESRP2 by siRNA directed against ESRP2 was judged from nuclear extracts. All but two sample sets from 16HBE14o-showed very low or absent ESRP2 protein signals and were excluded from quantitative analysis. (C) SCNN1B wild-type (exon3–exon4, see Fig. 1C, amplicon 4) and read-through alternative transcript (see Fig. 1C, amplicon 1) were visualized from cDNA derived from control and siRNA-treated cells. Data was evaluated based on the threshold cycle Ct provided for an SCNN1B amplicon in comparison to the housekeeping gene aldolase from technical duplicates for each qPCR reaction. Independent experiments derived from 12 cell culture plates, three each for the conditions growth control (no Dharmafect or siRNA), transfection control (Dharmafect only), siRNA ESRP2 and siRNA scr. were included into the analysis if at least two out of three samples from all conditions gave valid results for all three amplicons (n = 3 for T84 and n = 3 for 16HBE14o). All samples for 16HBE14o 24 h after transfection had to be excluded as SCNN1B transcripts could not be detected in the majority of qPCR reactions. ΔCt values were calculated for each sample as Ct[SCNN1B]-Ct[aldolase]. To compare differential expression between two triplicate sets, ΔΔCt was calculated for all possible nine combinations comparing three case and three reference samples. To assess the influence of Dharmafect, ΔΔCt[Dharmafect] was calculated as ΔΔCt[Dharmafect] = ΔCt[growth control] – ΔCt[transfection control]. The influence of an siRNA ΔΔCt[siRNA ESRP2] and ΔΔCt[siRNA scr.] treatment was calculated as ΔΔCt[siRNA ESRP2] = {ΔCt[siRNA ESRP2] – ΔCt[transfection control] – {mean ΔΔCt[Dharmafect]} and ΔΔCt[siRNA scr.] = {ΔCt[siRNA scr.] – ΔCt[transfection control]} – {mean ΔΔCt[Dharmafect]}. ΔΔCt[siRNA ESRP2] and ΔΔCt[siRNA scr.]. qPCR Data is shown for three independent experiments in each cell line (N = 3) whereby values derived from one biological replicate are displayed using the same color in black, grey or white circles, respectively. p values were calculated by Wilcoxon signed-rank test to test against the hypothesis that equal proportions of samples show an increase or decrease of SCNN1B transcript species upon treatment with siRNA. Only unrelated values—i.e. ΔΔCt derived from unshared ΔCt values within biological replicates—were included into the rank test. Significance in this test indicates a systematic effect of siRNA on SCNN1B transcription.