Figure 8.
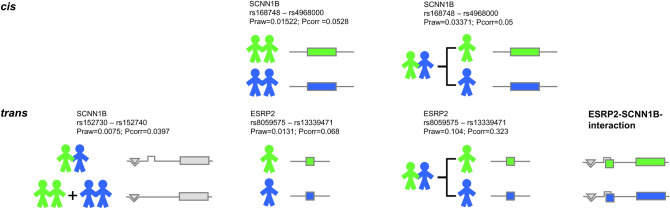
Identifying cis and trans regulatory elements in SCNN1B. Concordant mildly (two siblings who are shown in green), concordant severely (two siblings who are shown in blue) and discordant siblings (pairs composed of one green and one blue colored sib) have been selected from a total of 318 sibling pairs12 and analyzed in a series of case-reference association studies. Cis: The genomic segment rs168748–rs4968000 determines SCNN1B functionality directly: the observed interpair and matching intrapair association with CF disease severity can be explained by the SCNN1B variants in cis (data not shown). Trans: To explain the association with the discordant phenotype, variants at SCNN1B genomic segment rs152730–rs152740 require an interacting partner encoded in trans to SCNN1B. The genetic variation at ESRP2 contributes to the manifestation of SCNN1B functionality. In this model, binding sites for DNA interacting partners are shown as a triangular groove (for interaction partners shaped as a triangle) and as a square groove (for interaction partners shaped as a square). In the context of this work, ESRP2 is represented by a square and fits into its binding side at the SCNN1B locus. As a consequence, the phenotype of a sib within a discordant pair is shaped by the interaction of ESRP2 and SCNN1B: the allele rs152731-T, associated with intrapair discordance (Tables 1,2), is more vulnerable to ESRP2 (Fig. 6) and its genetic variations (Fig. 5). Functionally divergent ESRP2 alleles, visualized as green and blue squares, thus determine the phenotype of carriers with ESRP2-receptive SCNN1B alleles that are shown with a square groove. These ESRP2-receptive SCNN1B alleles include rs152731-T and are overrepresented among discordant, but rare among concordant pairs. The five displayed association signals correspond to: SCNN1B, rs168748–rs4968000, Praw = 0.01522; Pcorr = 0.0528: interpair comparison of allele distribution between 11 concordant mildly and 10 concordant severely affected sibling pairs (data not shown). SCNN1B, rs168748–rs4968000, Praw = 0.03371; Pcorr = 0.05: intrapair comparison of allele distribution between mildly affected siblings and severely affected siblings from discordant pairs (data not shown). SCNN1B, rs152730–rs152740, Praw = 0.0075; Pcorr = 0.0397: interpair comparison of allele distribution between 14 discordant sibling pairs and 21 concordant sibling pairs (see Fig. 1B). ESRP2, rs8059575–rs13339471, Praw = 0.0131; Pcorr = 0.068: comparison of allele distribution of unrelated patients stratified for the response of the nasal epithelium upon superfusion with amiloride assessed by nasal potential difference measurement (see Fig. 5). ESRP2, rs8059575–rs13339471, Praw = 0.104; Pcorr = 0.323: intrapair comparison of allele distribution between mildly affected siblings and severely affected siblings from discordant pairs.