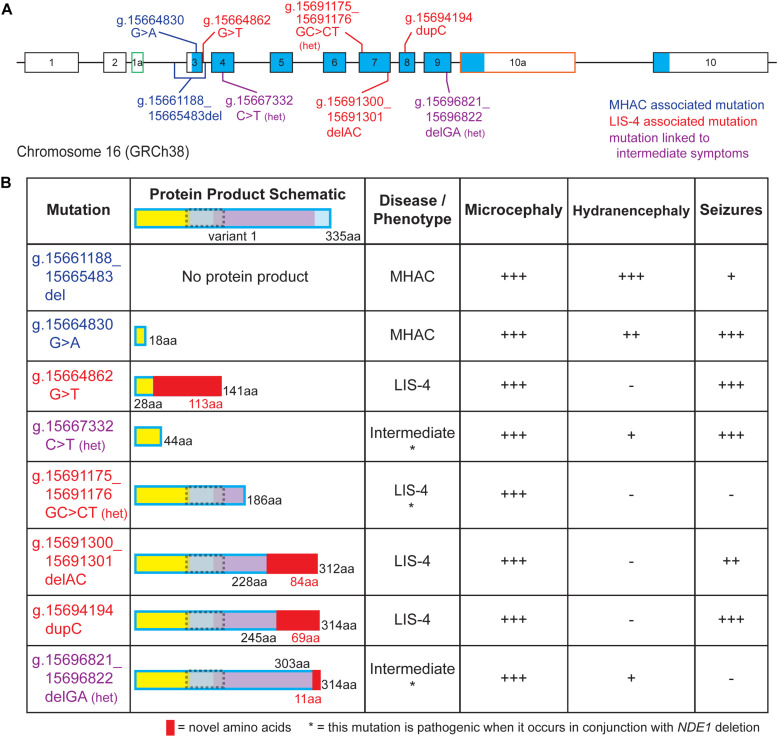
FIGURE 3.
Pathological mutations in NDE1 are linked to one of three clinical profile categories. (A) Schematic of human NDE1, including exons from isoforms 1, 2, and X2. Exons outlined in black are found in the canonical isoform (isoform 1), whereas alternative exons have either green or orange outlines. White fill indicates an untranslated region, whereas blue fill indicates the open reading frame. Mutations are color-coded by associated clinical profile type (blue = microhydranencephaly, or MHAC; red = lissencephaly-4, or LIS-4; and purple = intermediate). (B) Table of NDE1 mutations from (A), schematics of their expected protein product, their associated disease type, and presence and severity of microcephaly, hydranencephaly, and seizures in patients carrying the mutation. Protein schematics are color-coded by self-associated domain (yellow), LIS1-interacting domain (gray with dotted gray outline), NUDE_C domain (purple), sequence unassigned to a functional domain (blue), or novel sequence in the mutant protein (red). “Het” or * notes compound heterozygous mutations that are pathogenic when occurring alongside microdeletion of the NDE1 locus. All other mutations are pathogenic when homozygous. Symptom severity is noted as follows: –, not reported; +, mild; ++, moderate; +++, severe.