Fig. 1. nano-ChIA platform.
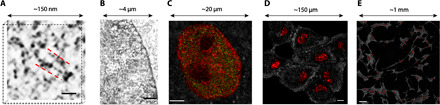
(A) ChromSTEM HAADF tomography characterizes the 3D chromatin structure of an A549 cell (contrast inverted). The inverted image contrast is inversely proportional to the local DNA density: As the electrons encounter a higher density of DNA along their trajectory, the image contrast appears darker. Individual nucleosomes and linker DNA are resolved at 2-nm spatial resolution. Scale bar, 30 nm. (B) ChromTEM imaging of a BJ cell nucleus on a 50-nm resin section prepared by ChromEM staining. Similar to ChromSTEM, ChromTEM also maps the DNA distribution, but the image contrast follows Beer’s law. Scale bar, 1 μm. (C) Coregistered PWS and STORM imaging of chromatin packing scaling (D, red pseudocolor) and active RNA Polymerase-II (RNAP II) (green) of an M248 cell nucleus. Scale bar, 3 μm. (D and E) Label-free PWS images of live A549 cells of both one field of view where chromatin packing variations within nuclei are visible (D) (scale bar, 20 μm) and a 9 × 9, stitched together, image to demonstrate the ability of PWS to visualize chromatin packing structure of cell populations (E) (scale bar, 100 μm). The pseudocolor represents the chromatin packing scaling inside the cell nuclei.
