Fig. 5. Proposed structure-based catalytic mechanism of Br2DNiR.
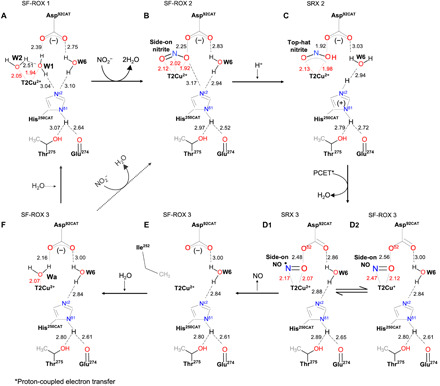
Solid arrows show the reaction sequence based on the structures of as-isolated and ligand-bound species of Br2DNiR described here (A to F). The further reaction of species (F) in CuNiRs that have a single water ligand of T2Cu in the resting state is shown as a dashed line. (A) Two waters bound in as-isolated SF-ROX structure 1 and in conformation 1 of as-isolated SRX structure 1. (B) Side-on nitrite bound in nitrite-bound SF-ROX structure 2. (C) Intermediate state with top-hat nitrite bound in nitrite-bound SRX structure 2. (D1) and (D2) represent two slightly different NO bound conformations in the SF-ROX 3 (D2) and SRX 3 (D1) structures; in D1, N and O of NO are almost equidistant. (E) Tricoordinate T2Cu site in NO-bound SF-ROX structure 3 where NO is no longer at the T2Cu site, resulting in the flipping of Ile252 into the space vacated from loss of fourth ligand. (F) Single water bound in NO-bound SF-ROX structure 3 where the water has been captured by the T2Cu site following the release of NO. The step from (F) to (B) represents the well-studied CuNiRs, such as AcNiR, AfNiR, and AxNiR, while (F) to (A) represents the return of Br2DNiR to its resting state.
