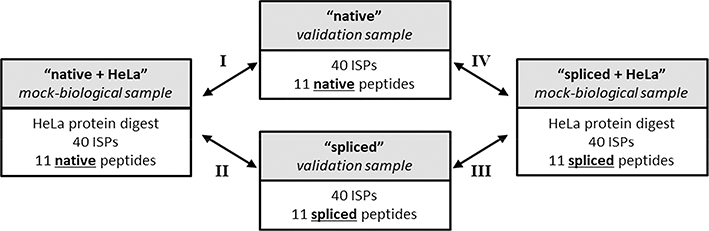
Figure 2.
Experimental design for benchmarking the P-VIS workflow. Eleven spectra for which Liepe et al. proposed spliced peptide matches and Mylonas et al. proposed native (non-spliced) matches were chosen. Synthetic peptides were obtained for each of these proposed sequences. All 11 spliced peptides were pooled together into a single sample, and all 11 native peptides were pooled together into a single sample. For each of these two samples, two aliquots were made, and HeLa cell protein extract was added to one of the aliquots to mimic the complexity of a biological sample. A mixture of 40 internal standard peptides (ISPs) was added to each of the four samples. The four samples were then analyzed to be either nano-flow or micro-flow LC–MS/MS. Arrows indicate the four pairwise data comparisons that were performed using PSM_validator. The labels for these comparisons (roman numerals I–IV) are referenced throughout the remainder of the manuscript.