FIGURE 2.
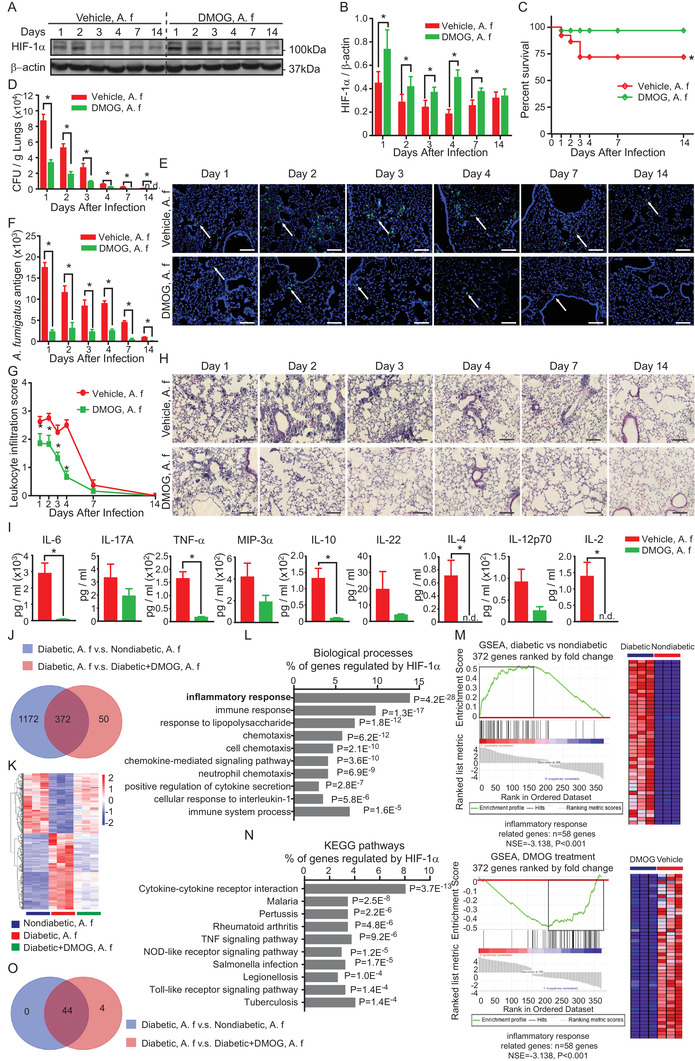
HIF‐1α induction attenuates the pulmonary infection and the inflammatory responses after pulmonary A. fumigatus infection in diabetic mice. Diabetic mice were injected with DMOG (300 mg/kg, i.p.) or vehicle every other day for 1 week before and 2 weeks after inoculation of 5 × 108 A. fumigatus conidia i.t. (A. f). A, Representative images of HIF‐1α and β‐actin western blots. B, Quantification of HIF‐1α protein expression normalized to β‐actin (n = 5). *P < .05 analyzed for each day using paired Student's t‐test. C, Survival rate of vehicle‐ or DMOG‐treated diabetic mice with pulmonary A. fumigatus infection (log‐rank test). D, Colony forming unit (CFU) counts per gram of lung tissue on indicated days post A. fumigatus challenge. N.d. denotes A. fumigatus were not detected (n = 5). E, Representative images of immunofluorescent staining of A. fumigatus (green) and DAPI (blue) in lung sections on indicated days post‐inoculation. Scale bars = 100 µm. White arrows indicate positive staining of A. fumigatus conidia and hyphae. F, Quantification of the green fluorescence intensity of A. fumigatus (n = 5). D and F, *P < .05 analyzed using unpaired Student's t‐test. G, Quantification of leukocyte infiltration (n = 5). *P < .05 compared between nondiabetic and diabetic groups analyzed using two‐way ANOVA followed by Bonferroni post‐hoc test. H, Representative images of H&E staining of lungs harvested on indicated days post‐A. f inoculation. Scale bars = 100µm. I, Cytokine levels in serum 2 days after A. fumigatus inoculation (n = 3 ‐ 5). *P < .05 analyzed using unpaired Student's t‐test. B‐I, Data are shown as mean ± SEM. J‐O, RNA purified from lung tissues harvested 2 days after A. fumigatus inoculation was used for global transcriptome analysis (n = 3). J, Venn diagram showing the number of deferentially expressed genes (log2 fold change > 2 or < −2, P < .01) in different groups, among which 372 genes that were regulated by DMOG were also differentially expressed in diabetes after A. fumigatus infection. K, Heatmap diagram showing the relative expression of the 372 genes that were dysregulated by diabetes and reversed by DMOG treatment. L and N, The top 10 gene ontology biological processes and KEGG pathway in which the 372 genes are involved. P‐values were determined by Fisher's exact test. M, Gene set enrichment analysis (GSEA) showing the enrichment of the genes involved in the inflammatory responses that were affected by diabetes (upper panel) and by DMOG (lower panel). O, venn diagrams showing the leading‐edge subset genes in panel M
Abbreviation: NES, normalized enrichment score.
