Fig. 1.
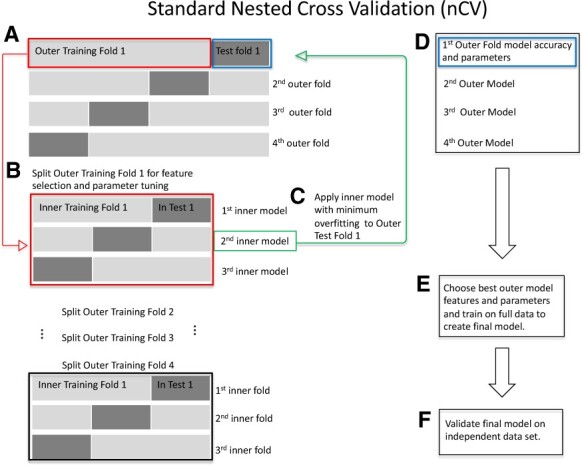
Standard nCV. (A) Split the data into outer folds of training and testing data pairs (four outer folds in this illustration). Then do the following for each outer training fold {illustration starting with Outer Training Fold 1 [red box (A)]}. (B) Split outer training fold into inner folds for feature selection and possible hyperparameter tuning by grid search. (C) Use the best inner training model including features and parameters (second inner model, green box, for illustration) based on minimum overfitting (difference between training and test accuracies) in the inner folds to test on the outer test fold (green arrow to blue box, Test Fold 1). (D) Save the best model for this outer fold including the features and test accuracies. Repeat (B)–(D) for the remaining outer folds. (E) Choose the best outer model with its features based on minimum overfitting. Train on the full data to create the final model. (F) Validate the final model on independent data. (Color version of this figure is available at Bioinformatics online.)
