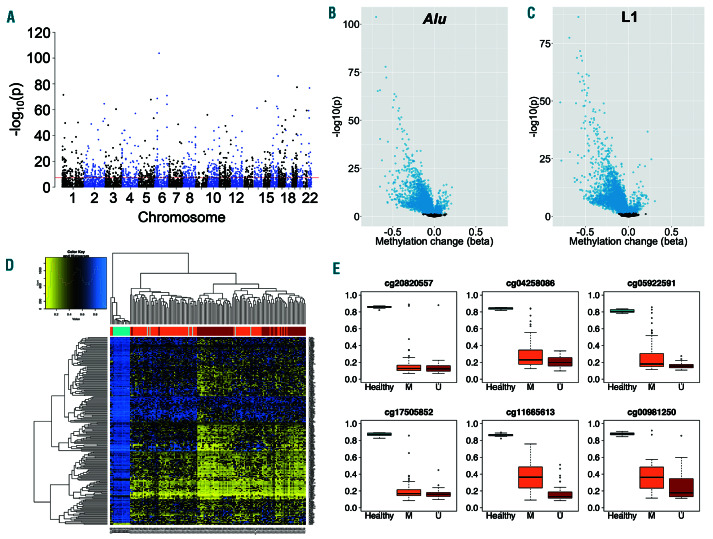
Figure 4.
Locus-specific differential methylation of retrotransposon elements in chronic lymphocytic leukemia. (A) Manhattan plot of retrotransposon loci showing differential methylation between chronic lymphocytic leukemia (CLL) patients (n=139) and normal CD19+ B cells from healthy individuals (n=14). The threshold (red line) indicates P<5x10-8. (B-C) Volcano plots for probes mapping to Alu (B) and L1 (C) elements. The change in methylation (Δb) and P-value (-log10(p)) are displayed, with significantly different loci (PFDR<0.05) highlighted in blue. (D) Heatmap displaying unsupervised clustering of the 200 most significantly differentially methylated loci. The colour scale indicates methylation level, from low (yellow) to high (blue). Healthy individuals (turquoise, n=14) and CLL patients with mutated (orange, n=75), unmutated (dark red, n=57) or unknown (grey, n=6) IGHV status are indicated. (E) Methylation of the six most significantly differentially methylated loci in healthy individuals (turquoise, n=14) and CLL patients with IGHV mutated (orange, n=75) and unmutated (dark red, n=57) disease. Lines indicate median values, boxes the interquartile range (IQR), whiskers the highest and lowest values within 1.5*IQR, and outliers are displayed as individual points.