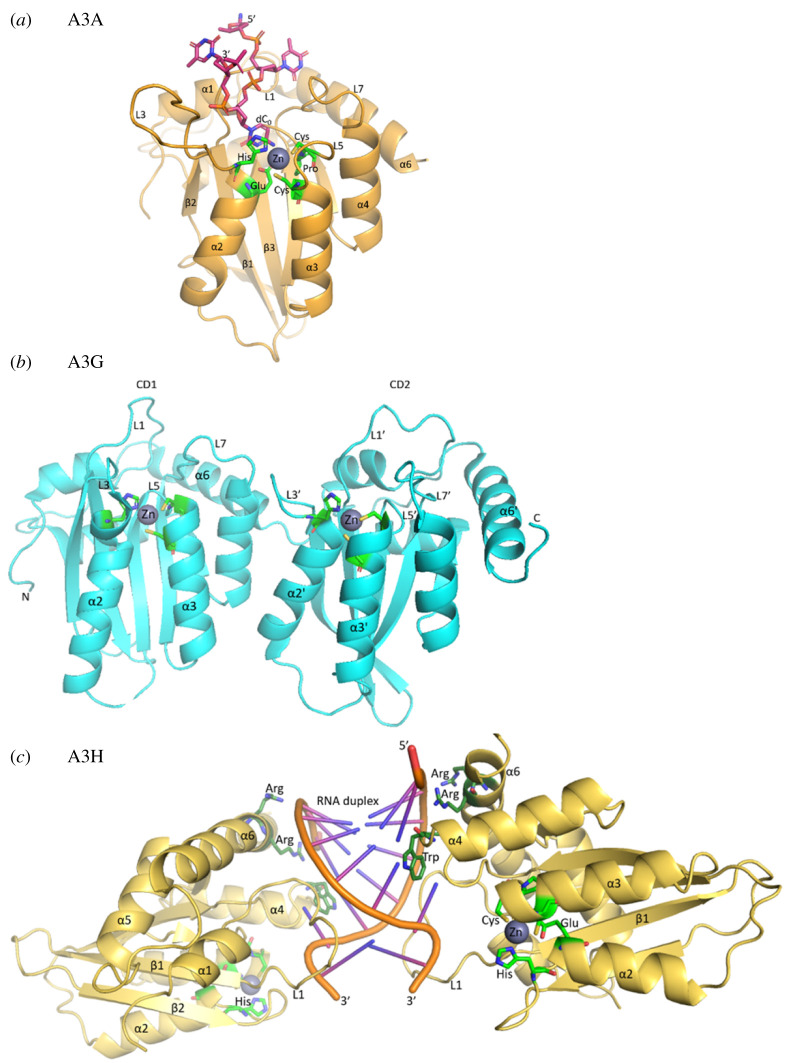
Figure 1.
A3 structures. (a) The core domain and active site of A3s, showing the single-domain A3A as an example. PDB 5KEG shown with modification to include the catalytic glutamate on α-helix 2 (α2) instead of alanine [70]. The consensus sequence His-X-Glu-X23–28-Pro-Cys-X2–4-Cys is represented as green residues coordinating a Zn atom. Pink carbon atoms represent the four nucleotide DNA (5′- dT2, dT1, dC0, dT-1 -3′) substrate. Blue, nitrogen; red, oxygen; yellow, sulfur and orange, phosphorus. (b) The double-domain A3G (PDB: 6P40) with loop 7' (L7') being the principal determinant of sequence specificity in the C-terminal domain (CD2) and loop 7 (L7) being a principle determinant for processivity and oligomerization (not shown) in the N-terminal domain (CD1) [71]. In both the CD1 and CD2, loops 1 (L1, L1'), 3 (L3, L3') and 5 (L5, L5') also contribute to DNA binding. (c) A3H RNA-mediated dimer (PDB 5W3 V) [72]. The catalytic residues (light green) coordinate the zinc atom. Key residues lying on α-helix 6 (α6) and loop 7 (L7) electrostatically mediate the protein DNA interface (dark green). The highlighted Trp residue lies on L7 forming a critical stacking interaction with nucleotide 3 of the RNA duplex.