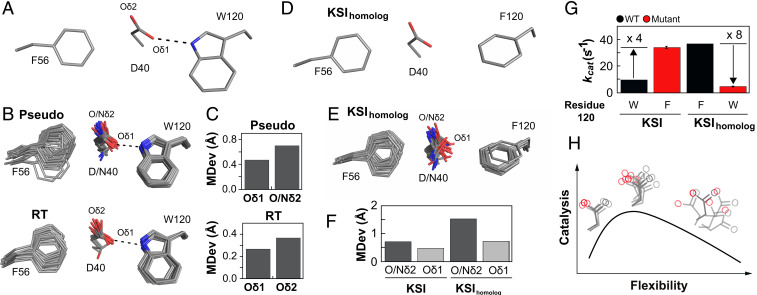
Fig. 6.
Ensemble–function analysis of general base catalysis for two KSI homologs. (A) Anion–aromatic and hydrogen-bonding interactions with F56 and W120 side chains, respectively, implicated in general base positioning in KSI (illustrated using PDB ID code 1OH0). (B) The full pseudoensemble (Upper) and RT ensemble (Lower) of the general base (D40), F56, and W120 side chains. (C) MDevs of the noncatalytic (Oδ1) and catalytic Oδ2 (Nδ2 when asparagine at position 40) oxygen atoms of the general base from the full pseudo- (Upper) and the RT ensemble (Lower). (D) Anion–aromatic interactions with F56 and F120 side chains implicated in KSIhomolog general base positioning (illustrated using PDB ID code 1OHP, KSI numbering used for KSIhomolog). (E) The KSIhomolog pseudoensemble (SI Appendix, Table S21). (F) MDev values for the D40 catalytic Oδ2 (dark gray bars, including asparagine Nδ2) and noncatalytic Oδ1 (light gray bars) obtained from the pseudoensembles in B and E. (G) Comparison of activity for KSIs with tryptophan and phenylalanine at position 120 (data from SI Appendix, Table S58). (H) Conformational ensemble model for optimal general base catalysis. Optimal flexibility enables the general base to shuffle protons between different positions in different substrates (Center); reducing motion (Left, e.g., when F120 in KSIhomolog is replaced with W120 in KSI) reduces catalysis, as does increasing motion (Right, e.g., from mutations in the loop carrying the general base) (53, 54).