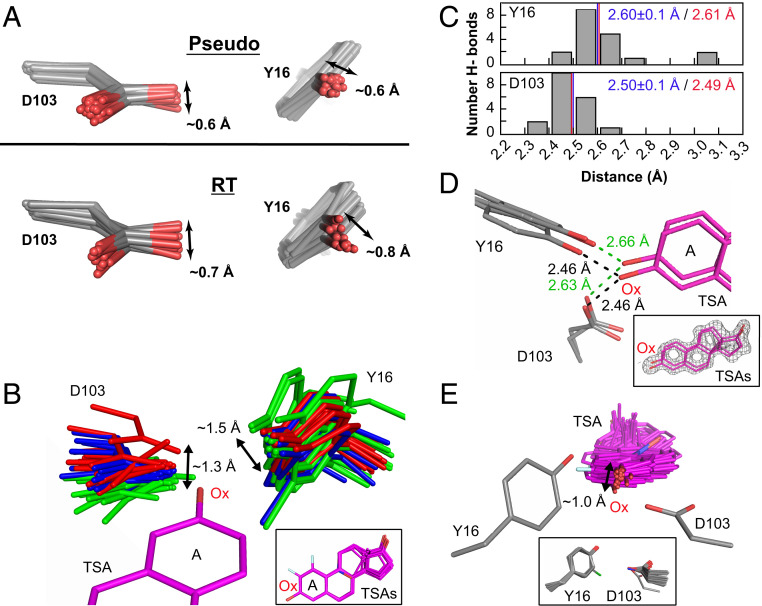
Fig. 7.
The KSI oxyanion–oxyanion hole conformational ensemble. (A) The oxyanion hole (Y16 and D103) pseudoensemble (Upper) and RT ensembles (Lower). Phenylalanine residues at position 16 are omitted, and chlorine atoms in chemically modified tyrosine residues have been omitted for clarity (see also SI Appendix, Figs. S16 and S23). (B) The KSI oxyanion hole as “seen” by 36 bound TSAs (equilenin and phenols) in cryo crystal structures (SI Appendix, Table S2). Structures have been color-coded in three groups according to the D103 position in space relative to the TSA oxyanion (Ox). All TSAs have been aligned on the A ring with only PDB 1OH0 shown for clarity. The Inset shows the 36 aligned TSAs. (C) Distribution of Y16 (Upper) and D103 (Lower) hydrogen-bond distances from an ensemble of KSI crystal structures of variants with WT-like activity and with a bound TSA (equilenin, n = 19) (SI Appendix, Table S2). The mean hydrogen-bond lengths and SDs from the cryo crystal structure distances are shown in blue and the distances obtained by solution 1H NMR are show in red (see SI Appendix, Fig. S25 for 1H NMR spectrum). (D) The TSA-bound RT multiconformer model shows that oxyanion hole Y16 and D103 and the bound TSA can make hydrogen bonds from different orientations. While multiconformer models do not allow unambiguous identification of each hydrogen bonded substates, possible hydrogen-bond lengths between Y16/D103 and the TSA are within the range obtained by cryostructures and solution 1H NMR. The Inset shows the TSA (purple sticks) and the experimental electron density (gray mesh, contoured at 1σ). (E) The bound TSAs from B as “seen” by the hydrogen bonding oxygens of Y16 and D103. Y16 and D103 have been aligned such that their hydrogen-bonding oxygens overlay but only one Y16/D103 set (PDB ID code 1OH0) is shown for clarity. The Inset shows all aligned Y16 and D103 hydrogen-bonding groups.