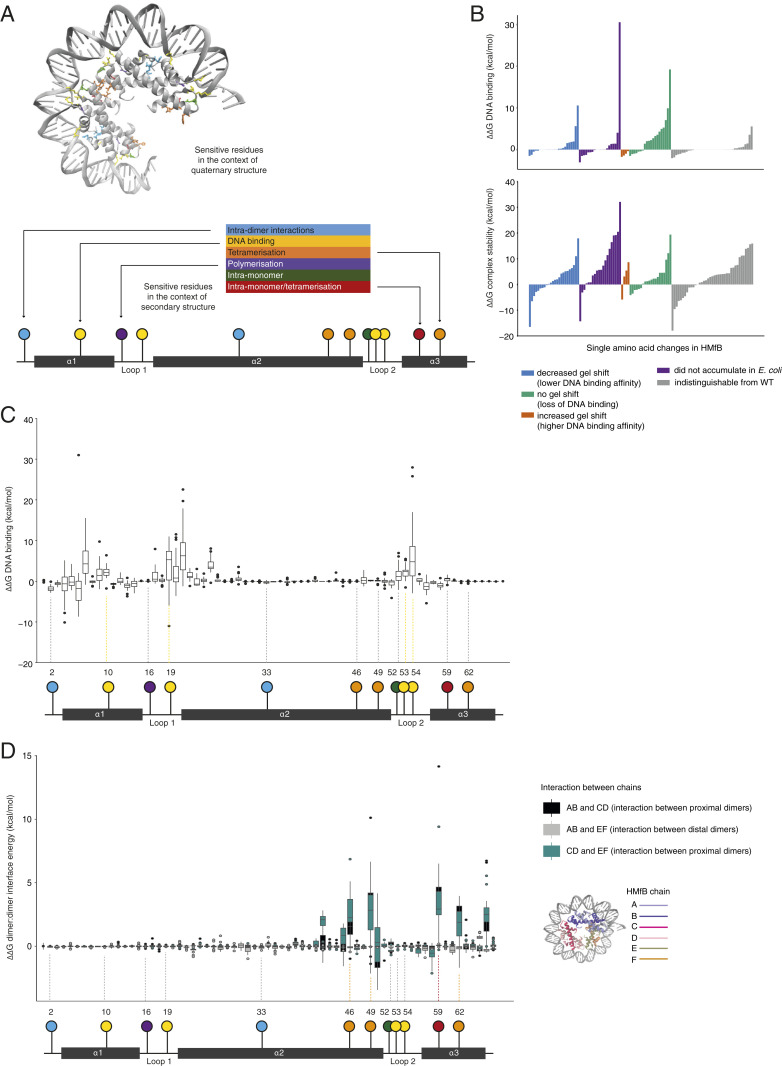
Fig. 6.
Modeling the impact of single amino acid mutations on the (HMfB)6–DNA complex. (A) Residues where mutations are known from previous experimental work (3, 18, 28–30) to affect monomer:monomer interactions, DNA binding, tetramerization, polymerization, and intramonomer interactions are highlighted on the quaternary and secondary structures. (B) FoldX-calculated changes in DNA binding affinity (Top) and stability (Bottom) for HMfB single amino acid mutants previously characterized qualitatively in gel shift experiments (30). Individual mutations are listed in SI Appendix, Table S2. (C and D) DNA binding (C) and tetramerization strength (D) for all possible single amino acid mutations of HMfB. The location of residues with previously known function is shown on the secondary structure beneath. For D, the resulting interaction energy between each dimer pair in the hexamer was calculated and the location of dimer pairs in the hexamer is shown. ΔΔG is quoted relative to the wild-type HMfB structure for all plots.