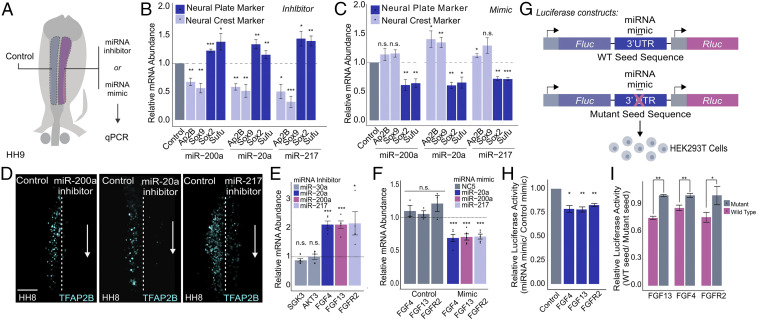
Fig. 4.
Neural crest miRNAs are required for specification and target components of the FGF signaling pathway. (A) Electroporation scheme for miRNA loss- or gain-of-function assays in which control reagent (blue) and targeted reagent (pink) were injected in different sides of a HH4 chicken embryo, which was then developed to HH8 to 9. (B and C) Quantitative RT-PCR for neural crest genes TFAP2B and SOX9 and neural plate genes SOX2 and SUFU upon miRNA loss (B) and gain of function (C) for the top three FGF-targeting neural crest miRNAs (miR-200a, miR-20a, and miR-217) (n = 6). (D) Immunohistochemistry for specification marker TFAP2B upon individually inhibiting FGF targeting miRNAs, miR-200a, miR-20a, and miR-217. Dotted line represents embryo midline. (E and F) Quantitative RT-PCR for target genes upon miRNA loss (E) and gain of function (F). miR-30a targets were excluded from gain-of-function assay due to a lack of observable changes in target gene expression upon loss of function. Gain-of-function assays were also performed using a nontargeting control (NC5) that does not affect target gene expression. (G) Diagram showing the experimental design for 3′ UTR luciferase reporter assays. Wild-type or mutant 3′ UTRs of target genes were cloned into a dual-luciferase reporter (pmiRGlo) and transfected into HEK293T cells with the corresponding miRNA mimic. For mutant 3′ UTRs, the miRNA mimic should not be able to bind the corresponding seed sequence. (H) Luciferase activity of wild-type 3′ UTR reporters. Each construct was transfected with the appropriate targeting miRNA mimic and normalized to a nontargeting miRNA mimic control (n = 3). (I) Luciferase activity of wild-type 3′ UTR luciferase reporters normalized to mutant 3′ UTR luciferase reporters, which should have no regulatory effect (n = 4). Error bars in B, C, E, F, H, and I represent the SD. HH, Hamburger and Hamilton; n.s., not significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Scale bar, 100 μM in (D).