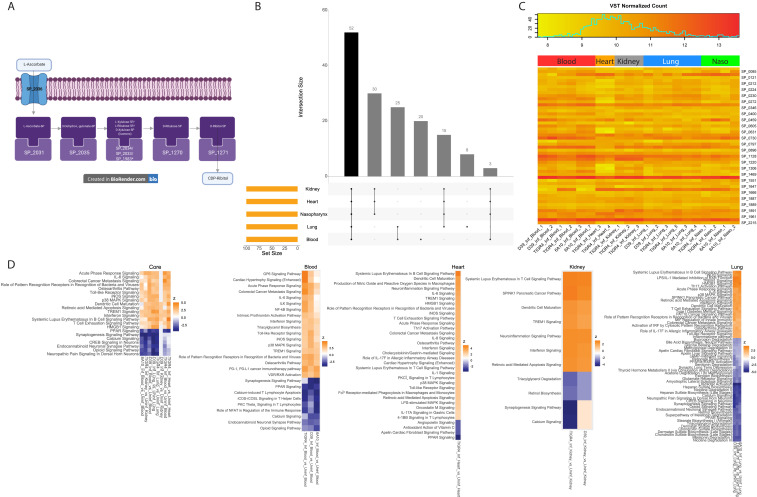
Fig. 5.
Pneumococcal and host genes of particular interest. (A) Schematic view of two potentially interlinked pneumococcal KEGG pathways (DatasetS6): Ascorbate and aldarate metabolism, and pentose and glucuronate interconversions. *SP_1983 was the only gene not DE relative to the nasopharynx. (B) Upset plot (see description in Fig. 4A) of the 100 most highly expressed Spn genes (Dataset S6) at each anatomical site (on average). (C) Absolute gene-expression level heatmap of the 52 Spn genes (Dataset S6) that are highly expressed genes across all sites (black vertical bar from B). (D) IPA activation z-score heatmaps (average z-score <−3 or >3) of canonical pathways in host DE genes (Dataset S7) present in all diseased anatomical sites. Nasopharyngeal pathways are not shown as too few host DE genes were observed (Fig. 4C).