Significance
The magnitude of CO2 efflux from soils (resulting from autotrophic and heterotrophic respiration) is one of the largest uncertainties in projecting future carbon–climate feedbacks. Despite research over several decades, the magnitude, direction, and duration of such feedbacks and their underlying microbial mechanisms are poorly understood, especially in the context of potentially interacting global environmental changes. In a decade-long experiment examining the interactive effects of CO2 and N enrichment, N limitation strengthened the stimulatory effects of elevated CO2 on soil respiration, primarily via N mining during the decomposition of more recalcitrant organic compounds. This study also provides a strategy for integrating genomics information into ecosystem and Earth system models to improve carbon-cycle predictions.
Keywords: elevated CO2, nitrogen deposition, soil respiration, metagenomics, Earth ecosystem model
Abstract
Whether and how CO2 and nitrogen (N) availability interact to influence carbon (C) cycling processes such as soil respiration remains a question of considerable uncertainty in projecting future C–climate feedbacks, which are strongly influenced by multiple global change drivers, including elevated atmospheric CO2 concentrations (eCO2) and increased N deposition. However, because decades of research on the responses of ecosystems to eCO2 and N enrichment have been done largely independently, their interactive effects on soil respiratory CO2 efflux remain unresolved. Here, we show that in a multifactor free-air CO2 enrichment experiment, BioCON (Biodiversity, CO2, and N deposition) in Minnesota, the positive response of soil respiration to eCO2 gradually strengthened at ambient (low) N supply but not enriched (high) N supply for the 12-y experimental period from 1998 to 2009. In contrast to earlier years, eCO2 stimulated soil respiration twice as much at low than at high N supply from 2006 to 2009. In parallel, microbial C degradation genes were significantly boosted by eCO2 at low but not high N supply. Incorporating those functional genes into a coupled C–N ecosystem model reduced model parameter uncertainty and improved the projections of the effects of different CO2 and N levels on soil respiration. If our observed results generalize to other ecosystems, they imply widely positive effects of eCO2 on soil respiration even in infertile systems.
Elevation of atmospheric CO2 concentrations, owing to fossil fuel combustion and land-use changes, represents one of the greatest scientific and political concerns of the 21st century (1). Carbon (C) movement into the atmosphere annually from soils (i.e., soil CO2 efflux or soil respiration) is much larger than annual C emissions from fossil fuel combustion (2), and thus even small changes in soil respiration could have significant impacts on the pace of change in atmospheric CO2. Numerous studies have demonstrated that elevated CO2 (eCO2) has a direct stimulatory effect on rates of plant photosynthesis (3), and an indirect positive effect on soil respiration, which typically includes autotrophic respiration from plant roots and heterotrophic respiration from microbial decomposition of litter and soil organic matter (SOM). The eCO2 stimulatory effect on soil respiration is commonly attributed to the following three mutually nonexclusive mechanisms from the actions of plants and microorganisms (4–7): enhanced root respiration associated with greater belowground plant biomass, enhanced microbial decomposition of fresh C due to greater supply of foliar and root-derived labile soil C, and increased microbial priming of old SOM fueled by this increased supply of labile soil C (4, 5). The stimulation of soil respiration by eCO2 (7, 8) has the potential to greatly accelerate the future rate of increase in atmospheric CO2 concentrations unless matched by an offsetting increase in net C uptake.
Human activities have also increased nitrogen (N) deposition to natural ecosystems (9). N enrichment is a growing concern because it disturbs N-cycle processes in many ecosystems (9). Various studies have suggested that N addition can either increase (10, 11) or reduce (12–15) soil CO2 efflux, while other studies have suggested that N addition does not influence soil CO2 efflux (16, 17), depending on ecosystem type and season of the year.
The stimulation of soil respiration by eCO2 also could be strongly influenced by variability in ambient soil N availability and the rate of atmospheric N deposition (18). However, studies that have explored the interactive effects of eCO2 and N on soil respiration are extremely scarce. For instance, an open-top study of young subtropical tree seedlings in contrasting eCO2 and N treatments in transplanted soil found that response to eCO2 was enhanced by high levels of N addition (10 g⋅m−2⋅y−1) in the earliest 2 y but unaffected by the same N supply in the subsequent year (19, 20). A free-air enrichment study in perennial grasslands also found no interaction between eCO2 and N addition treatments over the first 2 y of the study (21). Given that many questions about such potential interactions remain unresolved (22), here we report on 12 y of results in that same grassland study, assessing whether interactions develop and, if so, what underlying mechanisms might drive them.
It is well known that N availability alters many aspects of ecosystems (12, 23, 24) and thus could hypothetically influence responses of soil respiration to eCO2. Three potentially off-setting and interrelated mechanisms have been proposed. First, N limitation could affect belowground productivity and thus root respiration. For example, if N limitation constrains plant canopy development and the stimulatory effect of eCO2 on photosynthesis, and thus limits total productivity belowground, root respiration will decline (24). On the other hand, the same N limitation constraint on canopy development combined with stimulatory effects of eCO2 on photosynthesis could increase plant investment of C in nutrient-absorbing systems (25, 26), favoring C allocation to roots at the expense of aboveground biomass. Such a shift in allocation could increase root respiration (27). Second, changes in root detrital production and exudation of labile C into soils can influence substrate supply that fuels soil microbial activity and heterotrophic respiration. Third, the supply of labile C into soils can influence decomposition of SOM through the priming effect, which would also influence soil heterotrophic respiration (28). Under N limitation, greater photosynthesis caused by eCO2 could stimulate mining of N from SOM, and thus soil heterotrophic respiration, through enhanced priming mechanisms (29).
Although various studies indicate that N availability plays critical roles in mediating soil respiration (10–17, 23, 30, 31), divergent results are observed: positive (10, 11, 23), neutral (16, 17, 30), or negative (12–15, 30, 31). Thus, the impacts of N availability on the magnitude and duration of the eCO2 enhancement of soil respiration and its underlying mechanisms remain elusive, particularly under field settings. In addition, recent modeling efforts demonstrated the importance of understanding microbial C decomposition for more confidently extrapolating soil C cycling processes (32, 33). However, to date, it remains uncertain whether and how microbial processes influence the responses of terrestrial ecosystems to eCO2 and N deposition and how best to incorporate information regarding microbial responses to eCO2 and N into climate-C models for better simulation and prediction (32, 34, 35).
Herein, we report results from a well-replicated long-term (12 y at the time of sampling) CO2 × N experiment, BioCON (Biodiversity, CO2, and N deposition) (24), to elucidate the interactive effects of eCO2 and N enrichment on soil respiration and their underlying mechanisms. From 1998 to 2009, we measured soil CO2 efflux and other biogeochemical processes on 296 plots containing different numbers (1, 4, 9, or 16 species) and combinations (C3 and C4 grasses, forbs, and legumes) of perennial plant species at ambient CO2 (aCO2) or eCO2 (+180 ppm) with either ambient N supply (aN) or enriched N supply (eN, i.e., +4 g N⋅m−2⋅y−1). Hereafter, we refer to these four treatment combinations as aCO2-aN, eCO2-aN, aCO2-eN, and eCO2-eN. The contrasting high versus low levels of N supply in this study was a rough proxy for a part of the worldwide range of N supply rates in soils as well as for times or places with low versus high N deposition (24). Thus, we posit that the results are relevant to understanding the potentially different responses to eCO2 of both low versus high N fertility soils and contexts with low versus high N deposition. In 2009, we also assessed responses of microbial community functional gene structure to eCO2 and N enrichment to gain insights into microbial regulation of soil respiration. In addition, we incorporated microbial functional trait information into ecosystem models to explore means of better prediction of C cycling. Our overarching hypothesis is that N limitation would accelerate the stimulatory effects of eCO2 on soil respiration, primarily via microbial N mining mechanisms. We further explored the possibility that microbial functional trait information would greatly help to constrain the uncertainty of model parameters and hence significantly improve confidence in model simulations and predictions.
Results and Discussion
N Modulation of the Stimulatory Effect of eCO2 on Soil Respiration.
Soil CO2 efflux was measured ca. biweekly during the growing season (May to August) from 1998 to 2009. Overall, significantly (P < 0.01) higher soil respiration was observed at eCO2 than aCO2 at both low and high N supply (Fig. 1A), indicating that eCO2 stimulated soil respiration, consistent with previous reports (6, 7). Along with significant main effects of CO2, N, and plant species diversity as individual treatments, there were significant CO2 × N (P = 0.03; Table 1) and CO2 × N × year (P = 0.05) interactive effects on soil respiration, indicating that the stimulatory effect of eCO2 on soil respiration was modulated by N supply and that this interaction varied with time. Although the effect of eCO2 varied with plant diversity (P = 0.01 for the CO2 × plant diversity interaction; Table 1), the CO2 × N interaction was independent of plant diversity (P = 0.83 for the three-way interaction of CO2 × N × plant diversity; Table 1).
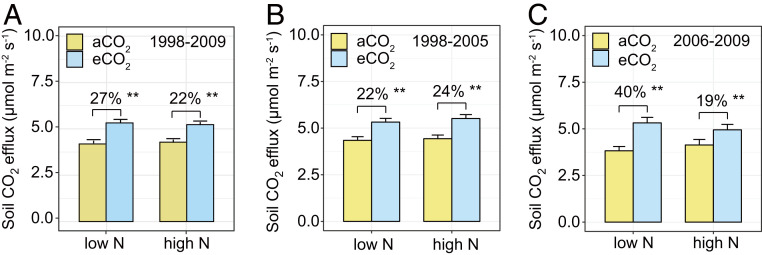
Fig. 1.
Observed responses of soil CO2 efflux to eCO2 at different N supply levels. (A) Soil CO2 efflux from 1998 to 2009. (B) Soil CO2 efflux from 1998 to 2005 (phase I). (C) Soil CO2 efflux from 2006 to 2009 (phase II). Each bar shows the annual mean plus SE of 74 plots. Percent changes of soil CO2 efflux in eCO2 plots relative to aCO2 plots are labeled above the bars. P values of the permutation t test are labeled **P < 0.01.
Table 1.
The main and interactive effects of CO2, N, and plant diversity (PD) on soil CO2 efflux measured from 1998 to 2009 based on repeated-measures mixed model across 296 plots
| F | P | |
| CO2 | 763.33 | <0.01 |
| N | 59.56 | <0.01 |
| PD | 692.89 | <0.01 |
| Year | 410.76 | <0.01 |
| CO2 × N | 4.63 | 0.03 |
| CO2 × PD | 13.99 | 0.01 |
| N × PD | 2.34 | 0.12 |
| CO2 × year | 9.02 | 0.01 |
| N × year | 15.69 | 0.01 |
| PD × year | 4.32 | 0.03 |
| CO2 × N × PD | 0.04 | 0.83 |
| CO2 × N × year | 3.73 | 0.05 |
| CO2 × PD × year | 3.02 | 0.08 |
| N × PD × year | 0.16 | 0.69 |
| CO2 × N × PD × year | 0.51 | 0.47 |
Significant (P < 0.05) effects are bolded.
To better identify the timing of the shift in the responses of soil respiration to eCO2 at contrasting N supplies, four commonly used change-point tests—Pettitt’s test, Buishand range test, Buishand U test, and standard normal homogeneity test (SI Appendix, Table S1)—were used. Our results indicated that 2005 was the breakpoint when the N influence on the stimulatory effects of eCO2 on soil respiration significantly changed (SI Appendix, Table S1). Therefore, we have divided the whole experimental period into two phases: phase I from 1998 to 2005 and phase II from 2006 to 2009 (see Materials and Methods for details). Using this breakpoint, the CO2 × N interactive effects on soil respiration significantly differed between these two phases, as indicated by a significant three-way interaction, CO2 × N × phase, on soil respiration (P = 0.02; SI Appendix, Table S2). In phase I, eCO2 significantly (P < 0.01) stimulated mean soil respiration regardless of N level (+22% vs. +24% at low and high N, respectively, Fig. 1B; P = 0.07 for the CO2 × N interaction, SI Appendix, Table S3). In contrast, the CO2 × N interaction became significant (P < 0.01; SI Appendix, Table S3) in phase II, and eCO2 stimulated mean soil respiration by 40% at low N supply but by only 19% at high N supply (Fig. 1C). These results indicate that long-term N limitation strengthened the stimulatory effects of eCO2 on soil respiration as the experiment proceeded.
Conceptually, the changing interactive effects of N and eCO2 on soil respiration between phase I and phase II were most likely due to soil processes, plant characteristics, and microbial community structure (21, 34, 36–40). Similar to soil respiration, significant (P < 0.01) CO2 × N × phase interactions were observed for soil net N mineralization rate and aboveground plant N concentration, but not for other soil and plant variables (SI Appendix, Table S2), indicating that there were temporal shifts in CO2 × N effects on those two variables. By examining the CO2 × N effect per year from 1998 to 2009, we found that the CO2 × N effect on soil respiration was significantly correlated with that on soil net N mineralization rate (P = 0.05), aboveground plant N concentration (P = 0.04), and aboveground plant C/N ratio (P = 0.03) (SI Appendix, Table S4). Further analysis revealed that eCO2 had no effect on net N mineralization rate at both N supplies in phase I but significantly increased the mineralization rate at high, but not low N supply, in phase II (SI Appendix, Fig. S1 A and B). In addition, aboveground plant N concentration was 8% lower at low than high N supply in phase I but was 20% lower in phase II (SI Appendix, Fig. S1 C and D). These data suggest that soil and plant N availability became more limited at low than high N supply as the time proceeded. The progressive N limitation could lead to less C allocation by plants to grow but more labile C inputs by eCO2 at low N supply (41), stimulating SOM decomposition and soil respiration. Collectively, the more positive soil respiration response to eCO2 at lower than higher N supply in phase II is probably related, at least in part, to the N-mediated phase shift of soil and plant N dynamics in response to eCO2. Similarly, microbes play important roles in regulating the interactive effects of CO2 and N on soil respiration, as discussed in the following section.
Roles of Microbial Processes.
The stimulation of soil respiration by eCO2 might be caused by changes in heterotrophic microbial processes and/or root-associated autotrophic processes (26). However, partitioning soil respiration into autotrophic and heterotrophic respiration is generally difficult (42). Thus, we used root biomass as a proxy to determine whether autotrophic respiration was a major component of our observed soil efflux interaction over time, given certain assumptions and caveats (43, 44). Root respiration is driven by a number of factors, including current soil temperature, prior soil temperature (which could drive acclimation), tissue N concentration, and soil water (45–48), as well as root biomass (43). Several of these factors (e.g., soil temperature, soil moisture, and root N concentration) showed no significant difference between eCO2 and aCO2 plots at both low and high N supply (SI Appendix, Table S5). Hence, although translating root biomass into absolute values of simulated soil respiration is challenging, assuming that root biomass is a reliable measure of relative differences in autotrophic respiration seems sound.
To evaluate whether root biomass mirrored the shifting N effect on eCO2 stimulation of soil respiration, we examined its responses to CO2 and N. In phase I, eCO2 stimulated root biomass to similar extents at low (11%) and high N (14%) supply (SI Appendix, Fig. S1E), which might partially account for the parallel responses of soil respiration to eCO2 at low and high N supply (Fig. 1B). In contrast, live root biomass was stimulated more by eCO2 at high N (22%) than low N (14%) supply in phase II (SI Appendix, Fig. S1F), whereas soil respiration was stimulated less by eCO2 at high N (19%) than at low N (40%) supply (Fig. 1C). Thus, live root biomass and associated autotrophic respiration responses likely were not the main drivers of the shifting responses of soil respiration to CO2 and N treatments, as mentioned above (SI Appendix, Table S4).
To examine the potential importance of different microbial processes in explaining the phase shift in CO2 × N interactive effects on soil respiration, we analyzed the composition and abundance of microbial functional genes for soil samples collected in 2009 using GeoChip (49). GeoChip is a generic microarray targeting hundreds of functional gene categories important to biogeochemical, ecological, and bioremediation processes. As predicted, the functional community structure was significantly shifted by CO2, N, and plant diversity treatments (SI Appendix, Table S6). All functional gene categories involved in C degradation and N cycling showed significant (P ≤ 0.05) or marginally significant (P ≤ 0.10) correlations across plots with mean soil CO2 efflux in phase II (SI Appendix, Table S7), but none of them did so in phase I (P > 0.10). Thus, microbial communities could play an important role in mediating the phase shift of N-induced differences in the soil respiration response to eCO2.
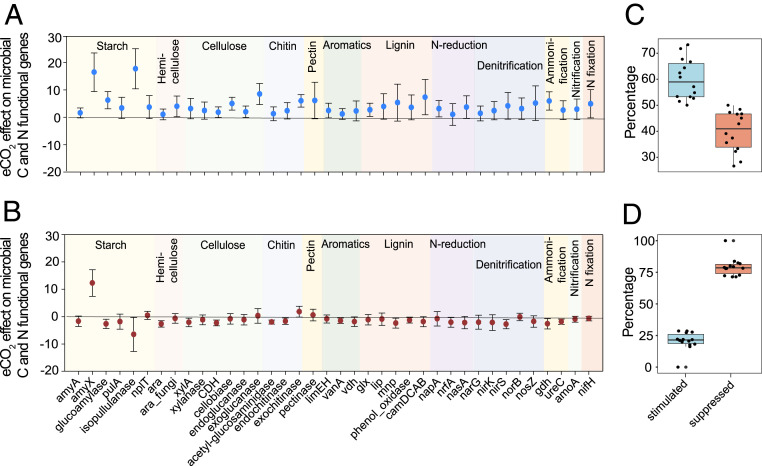
Directly relevant to questions of CO2 × N interactive effects on soil CO2 efflux in phase II, many microbial genes involved in C degradation and N cycling were significantly stimulated or suppressed by eCO2, but in different ways at low than at high N supply (Fig. 2). In general, at low N supply, most genes related to C degradation and N cycling were stimulated by eCO2 (Fig. 2A), whereas at high N supply most were slightly suppressed (Fig. 2B). Among those genes, antagonistic CO2 × N effects, whereby the combined CO2 and N effect on functional gene abundance was less than additive, were dominant (67%) (SI Appendix, Table S8), but no synergistic interactive effects were observed (50). Additionally, to summarize gene responses across all 14 assessed gene categories (in addition to those in Fig. 2 A and B), we determined the percentage of the significantly shifted genes (for each function) that increased versus decreased at eCO2 at each of the two N supply rates. A markedly greater percentage (59%) of affected genes were stimulated by eCO2 at low than at high N supply (Fig. 2C vs. Fig. 2D; P = 0.04 for CO2 × N effect on the relative abundance of those genes; SI Appendix, Table S6). Altogether, the changes in various functional gene abundances suggest enhanced microbial decomposition response to eCO2 at low N supply. These results are consistent with the above experimental observations that the effects of eCO2 on soil respiration in phase II were more enhanced at low N than at high N supply.
Fig. 2.
eCO2 effects on microbial functional genes important to C and N cycling at low and high N supply. Response ratios of functional genes at (A) low N supply and at (B) high N supply. Individual functional genes detected by GeoChip are shown on the x axis. Error bars indicate 95% confidence intervals of gene abundance difference between eCO2 and aCO2. (C) The percent of significantly shifted microbial gene probes stimulated (blue) versus suppressed (orange) by eCO2 at low N supply and (D) at high N supply. Percentages of stimulated and suppressed gene probes were averaged across gene probes in each gene category (each point in the boxplot) relevant to C, N, and P cycling. These gene categories (n = 14) include starch, hemicellulose, cellulose, chitin, pectin, aromatics and lignin degradation, N reduction, denitrification, ammonification, nitrification, N fixation, and phosphate limitation and phosphorus utilization.
In parallel with changes in overall community functions, CO2 and N showed antagonistically interactive effects on a variety of bacterial genes (26% of the bacterial genes on the arrays) related to C degradation and N cycling, which were significantly (P < 0.05) stimulated by eCO2 at low N supply but were suppressed by eCO2 at high N supply (SI Appendix, Table S9). However, only three fungal genes (15%) related to C degradation were antagonistically affected by CO2 and N, while most of the fungal genes (85%) showed similar responses to eCO2 at the two N supplies. The results suggest that high N supply suppressed the eCO2 effect on bacterial functional capacity, thus potentially shifting the microbial community toward relatively higher fungal capacity.
Two major competing, but nonexclusive, theories have been proposed to explain the mechanisms underlying the impacts of N on eCO2-induced microbial decomposition of SOM (23). Herein, we identify which ones may be at work in BioCON. The “stoichiometric decomposition” theory posits that microbial activity (e.g., decomposition and respiration) will be highest when the stoichiometry of substrates matches that of microbial demand and C and N colimit decomposition (51). Accordingly, soil respiration will be stimulated more by eCO2 at high than at low N supply (SI Appendix, Table S10). This is because with higher substrate C/N ratios at eCO2 and low N supply microbes are unable to meet their N demand, which may suppress microbial C decomposition rates and disfavor rapidly growing microbes (r-strategists) that primarily use labile C. In contrast, the “microbial N mining” theory asserts that, at low N availability, microbes use labile C as an energy source to decompose recalcitrant SOM to acquire N, accelerating microbial decomposition of SOM and favoring genes involved in recalcitrant C degradation (slow-growing k-strategists) (SI Appendix, Table S10) (52).
Data from BioCON in phase II are more consistent with the microbial N limitation and N mining theory. eCO2 significantly increased soil net N mineralization at high, but not low, N supply (SI Appendix, Fig. S1B) and the aboveground plant N concentration and total plant N pool were considerably less under low than high N supply (SI Appendix, Fig. S1 D and H). Those results suggest limited N availability at low N supply may not have met microbial N demand, and hence microbial C decomposition was stimulated to acquire N. As a likely result, most genes involved in C and N cycling were stimulated by eCO2 at low N supply (Fig. 2A), in contrast to their suppression by eCO2 at high N supply (Fig. 2B). Alternatively, eCO2 weakly (P = 0.08) decreased soil C/N ratio at low but not high N supply (SI Appendix, Fig. S1J). As microbial C content relative to N is one to two orders of magnitude lower than that of plants (51), a decreased soil substrate C/N ratio may relieve nutrient limitation and promote substrate-induced microbial respiration (53), echoing the stoichiometric decomposition theory. It should be noted that N addition could reduce soil respiration (12–15) by suppressing microbial decomposition via both N mining and substrate stoichiometry, which are time-dependent and may take a long time to appear. This could be one of the main reasons that the N-induced suppression of the stimulatory effects of eCO2 on soil respiration was more obvious in phase II.
Decomposition Modeling Enabled by Microbial Functional Traits.
As demonstrated above, microbial functional community structure likely plays an important role in mediating responses of soil respiration to eCO2 and N availability. Such information is a prerequisite for predicting how the soil microbial community and associated functions respond to multiple global change factors. The next urgent need is to translate such conceptual understanding into an ecosystem model-based quantitative framework because process-based microbial-explicit ecosystem models can provide mechanistic insights, integration, and scenario testing not available from or possible with experiments (54). In this regard, microbial-explicit ecosystem models will enable us to mechanistically simulate large-scale experiments that would be too costly to establish in reality and predict their future dynamics. However, a grand challenge in ecology is how to integrate microbial functional traits into ecosystem models to improve their performance and predictive ability (55).
To address the above challenge, we incorporated the GeoChip-detected microbial functional genes into the C–N coupled microbial-enzyme decomposition (MEND) model (SI Appendix, Fig. S2A and Tables S11–S15). We used tMEND to denote the MEND model parameterized with traditional observations such as soil CO2 efflux and mineral N concentrations. For comparison, gMEND refers to the MEND model calibrated with additional GeoChip-based microbial functional gene abundance data (Fig. 3A and SI Appendix, Fig. S3A). We compared the results of these two microbial models (tMEND, gMEND) plus a third model, the nonmicrobial C-only terrestrial ecosystem (TECO) model (SI Appendix, Fig. S2B). In addition to the best fit between observed and simulated soil CO2 efflux and mineral N (NH4+ and NO3–) concentrations, we constrained the model by achieving the highest goodness of fit between MEND-modeled relative changes in enzyme concentrations and GeoChip-detected relative changes in oxidative and hydrolytic gene abundances in response to eCO2 (SI Appendix, Table S11).
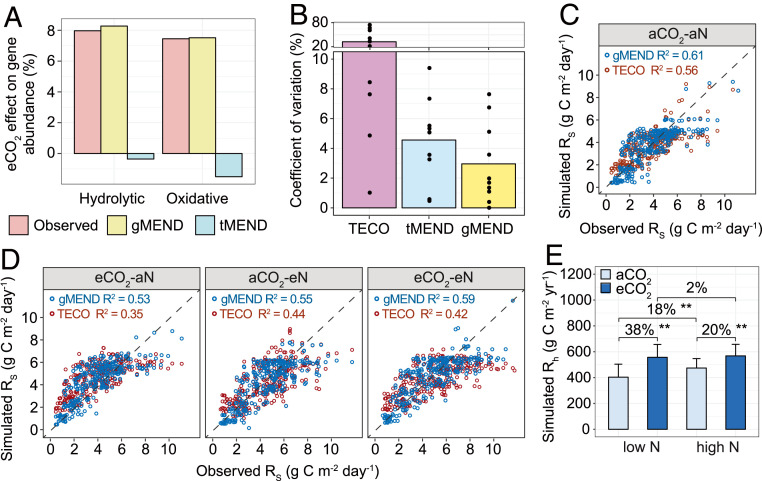
Fig. 3.
Model simulations. (A) Comparison of eCO2-induced percent changes of hydrolytic and oxidative enzymes observed by GeoChip to the simulated effects by gMEND and traditional MEND without gene information (tMEND) at low N supply. The GeoChip data were obtained from the samples from 2009. (B) Parameter uncertainty quantified by the coefficient of variation (CV) for the nonmicrobial C-only TECO, tMEND, and gMEND models; the bars show mean CV of 10 calibrated parameters represented by dots. (C) Model calibration with the soil respiration (Rs, 1998 to 2009) at aCO2-aN. (D) Model validations were performed using Rs at eCO2-aN, aCO2-eN, and eCO2-eN for gMEND and TECO. (E) Percent changes of gMEND-simulated heterotrophic respiration (Rh) between different CO2 and N levels. The error bars represent SEs. P values of the permutation t test are labeled as **P < 0.01.
The eCO2-induced changes in hydrolytic and oxidative genes observed by GeoChip were consistent with changes simulated by gMEND but not tMEND (Fig. 3A). Also, the parameter uncertainty (i.e., coefficient of variation) of gMEND was considerably reduced compared to both tMEND (by 35%) and the nonmicrobial C-only TECO model (by 86%; Fig. 3B). As a result, the gMEND model was able to simulate the observed soil CO2 efflux at aCO2-aN relatively well (R2 = 0.61; Fig. 3C). In addition, the gMEND model that had been calibrated only with the data at aCO2-aN was further validated against independent datasets from the other three CO2 and N treatments. The performance was almost as good as model calibration for ambient conditions (5% less variance explained on average) (R2 = 0.53 to 0.59; Fig. 3D). In contrast, the TECO model explained considerably less variation in observed soil respiration at the other three treatment combinations (R2 = 0.35 to 0.44; Fig. 3D) than at ambient conditions (explaining about 16% less of the variance). These differences suggest that gMEND better adjusts for CO2 and N effects than TECO. Finally, gMEND-simulated ammonium and nitrate concentrations also agreed fairly well with the observations (SI Appendix, Fig. S3B). Altogether, the above results suggested that the gMEND model can capture the dynamics of soil CO2 efflux reasonably well, comparable to or better than several previously field modeling studies (56, 57).
We further estimated eCO2-induced soil C loss via heterotrophic respiration. Our simulations showed that eCO2 would cause 38% and 20% more heterotrophic respiration at low and high N supply (Fig. 3E), respectively, and that enriched N would lead to 18% and 2% more heterotrophic respiration at aCO2 and eCO2 (Fig. 3E), respectively. We then asked what the implications might be if such results were general for grasslands globally. Applying our results to the world’s grasslands based on the International Geosphere-Biosphere Program classification scheme and the estimated annual soil respiration from grasslands between 2001 and 2009 (58), eCO2 (+180 ppm) alone would increase heterotrophic respiration by 1.6 ± 0.1 Pg C⋅y−1 whereas enriched N (+4 g N⋅m−2⋅y−1) alone would increase heterotrophic respiration by 0.8 ± 0.2 Pg C⋅y−1. However, combined eCO2 and enriched N would increase heterotrophic respiration by 1.7 ± 0.2 Pg C⋅y−1 across global grasslands, 29% less than the additive effects of eCO2 and enriched N alone. Thus, interactions noted herein could be significant globally.
Although our modeling results via calibration (Fig. 3 A–C) and validation (Fig. 3D) indicated that the gMEND could encapsulate the dynamics of soil CO2 efflux fairly well, about 40% of the variation was not captured, likely for two primary reasons. First, various experimental measurements such as gross primary productivity, soil CO2 efflux, temperature, moisture, and microbial traits were highly variable and some were uncertain, which could contribute to the discrepancy between model simulations and experimental observations. Second, the MEND model used in this study does not consider the differential roles of diverse microbial communities (e.g., bacteria and saprotrophic and mycorrhizal fungi) in regulating C–N cycling in response to eCO2 and enriched N supply owing to our poor understanding of these processes (8). Incorporating additional biological processes and their interactions into the MEND model may improve the modeling of soil CO2 efflux and its response to environmental change (8). Nevertheless, this study demonstrates the feasibility of integrating massive omics information into ecosystem models for better predictions of the soil C response to eCO2 and enriched N.
Conclusions
We found that the positive effect of eCO2 on soil respiration at low N supply was greater in years 9 to 12 than in years 1 to 8 of a long-term experiment and that changes in microbial functional traits, such as functional genes involved in C and N cycling processes, as well as temporal shifts in soil and plant N availability, likely underlie this dynamic. These findings would, if general, have important implications for predicting the responses of ecosystems to future environmental changes. For example, considering that N limitation is widespread in natural ecosystems, considerable stimulation of soil respiration in response to rising CO2 concentration might occur. Pervasive N deposition due to anthropogenic activities could offset, at least partially, the stimulation of soil respiration by elevated atmospheric CO2, and thus could weaken the positive feedback between the terrestrial C cycle and climate change. Our study also shows that whether microbially mediated feedback to rising CO2 concentrations and climate change is positive or negative depends on microbial functional groups and whether their associated functions are stimulated by eCO2, suggesting the necessity of integrating microbial functional traits into climate-C models for better prediction (34, 55). As expected, incorporating those functional genes into a coupled C–N ecosystem model substantially reduced model parameter uncertainty and improved the prediction of soil respiration in response to eCO2 and enriched N supply. Although further model development, calibration, and validation of a microbially enabled model will require rigorous benchmarking with observations, this study serves as a step forward to mechanistically assimilate microbial functional traits into climate-C cycle modeling.
Materials and Methods
Experimental Design and Sampling.
The BioCON experiment contains 296 main plots with a fully factorial 2 × 2 × 4 combinations of three treatments: CO2 (ambient vs. +180 ppm), N deposition (ambient vs. +4 g N⋅m−2⋅y−1), and plant diversity (1, 4, 9, or 16 species) (59). Plots were established with diversity treatments in 1997. The CO2 and N treatments began in 1998. The 296 plots are evenly distributed among six rings with split-plot arrangement of CO2 and N treatments. CO2 treatment is the whole-plot factor. The subplot N and plant diversity treatments were randomly distributed and replicated in individual plots among the six rings. Although ambient CO2 concentration has increased during the experimental period, resulting in inconstant ambient CO2 concentrations over time, a free-air CO2 enrichment system is used to provide a constant elevation of CO2 by an average of 180 ppm above ambient in three elevated CO2 (eCO2) rings. The other three ambient CO2 rings (aCO2) were treated identically but without additional CO2. Half of the plots in each ring received N amendments of 4 g N⋅m−2⋅y−1 applied as NH4NO3 on three dates each year. As a consequence, there were in total four CO2 and N treatments among 296 plots: aCO2 and low N (aCO2-aN), eCO2 and low N (eCO2-aN), aCO2 and high N (aCO2-eN), and eCO2 and high N (eCO2-eN), with each treatment having 74 plots (biological replicates). For each of the four CO2 and N treatments, there were 32 plots planted with 1 species, 15 plots planted with 4 species, 15 plots planted with 9 species, and 12 plots planted with 16 species (59).
Plant and Soil Variables.
Each year (1998 to 2009) in every plot, above- and belowground (0- to 20-cm depth) plant biomass were mainly measured in August (59). Soil net N mineralization rates were measured in situ each year in each plot for a ca. 1-mo period using a semiopen core in July (24). Net N mineralization is the net transformation of N from organic to inorganic forms and is considered to represent the availability of N to plants. Plant N concentration (percent aboveground plant and root) and plant C/N ratio (aboveground plant and root) were measured in August from 2001 to 2009. Soil C/N ratio was measured in years 2002 and 2007.
Soil CO2 efflux in each plot was measured for 11 to 36 times per year using a LI-COR 6400-09 soil CO2 efflux chamber from 1998 to 2009. Measurements made during peaking growing seasons (from May to August) were used in this study, as those data best reflect growing season ecosystem functioning. Within each of those months, soil CO2 efflux was measured two to five times in each plot. In the short term, soil CO2 efflux measured using chamber techniques may deviate from the instantaneous soil respiration due to changing CO2 stored in the soil pore space (60). However, in the medium to long term, soil CO2 efflux corresponds to soil respiration as all CO2 produced in the soil must be emitted from the soil. Thus, in this study, we use “soil CO2 efflux” and “soil respiration” in an exchangeable way.
GeoChip Experiments and Raw Data Processing.
Soil samples for microbial community analysis were collected from the 296 plots in August 2009. Microbial genomic DNA was extracted from 5 g of well-mixed soil for each sample by combining freeze-grinding and sodium dodecyl sulphate for cell lysis and purified by agarose gel electrophoresis, followed by phenol–chloroform–butanol extraction as previously described (61). The functional gene array GeoChip 4.0 was used for DNA microarray hybridization. As described previously (62), the DNA samples were labeled with fluorescent dye Cy-3 dUTP and hybridized with the slides with GeoChip 4.0M in a rotator/incubator at 67 °C plus 10% formamide and rotated at 20 rpm for 24 h. After hybridization, GeoChip was scanned at 100% laser power and 100% photomultiplier tubes gain with a NimbleGen MS 200 Microarray Scanner (Roche NimbleGen). Scanned images were gridded by NimbleScan software (Roche) to obtain the signal intensity for each probe. Raw data obtained from NimbleScan was submitted to the Microarray Data Manager at http://ieg.ou.edu/microarray/ and analyzed by the data analysis pipeline (49). We removed spots with the signal-to-noise ratio below 2, considered as poor quality.
Model Simulation and Prediction.
Details for modeling methods are provided in SI Appendix, Supplementary Information Text. Briefly, we used a nonmicrobial C-only TECO model and a C–N coupled MEND model to simulate daily soil CO2 efflux for four CO2 and N treatments from 1998 to 2009. In TECO model, we used a group of first-order ordinary differential equations to describe the C turnover among fast, slow, and passive SOM pools (SI Appendix, Fig. S2B). We set prior ranges of C turnover rates based on a previous study (63), which were modified by soil temperature (T) and moisture (W) during the simulations. In comparison, The C–N coupled MEND model describes both C and N transformation processes in the following pools: oxidative and hydrolytic particulate organic matter (POMO and POMH), mineral-associated organic matter (MOM), active MOM (QOM), dissolved organic matter (DOM), active and dormant microbial biomass (MBA and MBD), three enzyme functional groups, and mineral N (NH4+ and NO3–) (SI Appendix, Fig. S2A). The two POM pools are decomposed by oxidative or hydrolytic enzymes, while the MOM is decomposed by both. Model state variables, governing equations, component fluxes, and parameters are shown in SI Appendix, Tables S12–S15.
The modified shuffled complex evolution algorithm was used to calibrate model parameters for both TECO and MEND models under the aCO2-aN treatment (SI Appendix, Supplementary Information Text). We then validated the model using the same set of model parameters calibrated for aCO2-aN to simulate soil CO2 efflux under the other three treatments. Microbial gene abundances were used as objective functions to calibrate model parameters only for the gMEND model (57). The coefficient of determination (R2) was used to estimate the model performance between simulated and observed soil CO2 efflux (64). Additional observational variables (NH4+ and NO3– concentrations and response ratios of oxidative and hydrolytic enzymes) for MEND model calibration and validation are shown in SI Appendix, Table S11. Parameter uncertainty of TECO model was quantified by probabilistic inversion (Markov chain Monte Carlo) algorithm while that of MEND model was quantified by the critical objective function index method.
Statistical Analyses.
Since microbial community structure was determined with all 296 soil samples collected in 2009, this study focused on the soil CO2 efflux from the beginning of the BioCON experiment until 2009. To identify the year in which interaction between CO2 and N emerged, we calculated the response ratio (RR) of soil CO2 efflux differences between eCO2 and aCO2 at low or high N supply in every month of the growing season. The N influence was then calculated as RR at high N supply minus RR at low N supply, representing the CO2 × N interaction. The annual mean value of the N influence was calculated for each year. Four commonly used change-point tests, including Buishand range test, Buishand U test, standard normal homogeneity test, and Pettitt’s test, were performed on the annual mean values of the N influence. Because no soils were collected for microbial analysis in phase I, most of the statistics-based mechanistic analyses were focused on phase II.
For each year from 1998 to 2009, data points of soil CO2 efflux (micromoles per mole2 per second) that were higher than mean plus 1.96 SDs or lower than mean minus 1.96 SDs of all data points in a plot were regarded as outliers and removed before the analysis (65). By doing this, we reduced the within-plot variation in soil CO2 efflux measurements to enhance the statistic power. We used the same approach to identifying and excluding outliers for other soil and plant variables, including soil net N mineralization rate (milligrams per kilogram per day), soil temperature (degrees Celsius), soil moisture, soil pH, soil C/N ratio, plant N concentration (percent), plant C/N ratio, plant biomass (grams per meter2) and plant N pool (grams per meter2). Net N mineralization data in 2008 were contaminated and thus were not included in the analysis (41). The significance of CO2 × N effects and CO2 × N × phase effects on soil CO2 efflux, soil, and plant variables was tested using repeated-measures mixed models following the previous method (66). The CO2 × N effects (N influence on the eCO2 effect) on each of the soil and plant variables and on soil CO2 efflux were calculated per year from 1998 to 2009, then relationships between CO2 × N effects on soil/plant variables and on soil CO2 efflux were examined using Pearson correlation.
The eCO2 effects on soil and plant variables as well as microbial functional genes at low and high N supply were calculated based on Eqs. 1 and 2:
| [1] |
| [2] |
where , , , and represent mean of soil CO2 efflux, soil variables, plant variables, or the relative abundance of microbial functional genes in eCO2-eN, eCO2-aN, aCO2-eN, and aCO2-aN plots, respectively. Permutation t test was conducted to examine the significance of the eCO2 effect on plant and soil properties at both low and high N supply (67). At the low or high N supply, the significance of eCO2 effect on the abundance of each functional gene (total abundance of all probes of this gene; SI Appendix, Table S8) was examined by response ratio with 95% confidence intervals of gene abundance differences between eCO2 and aCO2 plots. We also examined the eCO2 effect on the abundance of each gene probe by response ratio. Of all significantly changed probes of an individual gene, we calculated the percentages of stimulated and suppressed probes by eCO2. Then, we calculated the averaged percentages of stimulated and suppressed probes across genes in different gene categories for C cycling, including starch, hemicellulose, cellulose, chitin, pectin, aromatics and lignin degradation, gene categories for N cycling, including assimilatory/dissimilatory N reduction, denitrification, ammonification, nitrification, and N fixation as well as gene categories for phosphorus (P) cycling, including P fixation and P utilization.
To determine the direction (additive, synergistic, or antagonistic) of interactive effects of CO2 and N on functional genes, we compared the observed effects (OEs, i.e., combined eCO2 and enriched N effects) and the expected effects (EEs), that is, additive effects of eCO2 alone and enriched N alone (50). For each functional gene, OE was calculated as follows: . EE was calculated as follows: + . The interactive effects are additive when OE is not different from EE. Interactive effects are synergistic if OE is significantly higher than EE or antagonistic if OE is significantly lower than EE. The significance of the interactive CO2 and N effect on each functional gene was tested by the permutational multivariate analysis of variance (Adonis) using the abundance matrix of this microbial functional gene.
Supplementary Material
Acknowledgments
The data analysis by Q.G. was supported by the National Science Foundation of China (41825016), the Second Tibetan Plateau Scientific Expedition and Research program (2019QZKK0503), and the special fund of the State Key Joint Laboratory of Environment Simulation and Pollution Control (19L01ESPC). The BioCON experiment was funded by the US Department of Agriculture (USDA) (Project 2007-35319-18305) through NSF-USDA Microbial Observatories Program, Long-Term Ecological Research (LTER) grants DEB-0620652, DEB-1234162, and DEB-1831944, Long-Term Research in Environmental Biology (LTREB) grants DEB-1242531 and DEB-1753859, Biological Integration Institutes grant NSF-DBI-2021898, Ecosystem Sciences grant DEB-1120064, and Biocomplexity grant DEB-0322057 and by US Department of Energy Programs for Ecosystem Research grant DE-FG02-96ER62291 and the University of Minnesota to P.B.R. and/or S.E.H. The experimental measurements with GeoChip were supported by the USDA (Project 2007-35319-18305) through the NSF-USDA Microbial Observatories Program, and the modeling work was supported by the US Department of Energy, Office of Science, Genomic Science Program under Awards DE-SC0004601, DE-SC0010715, DE-SC0014079, DE-SC0016247, and DE-SC0020163 and by the Office of the Vice President for Research at the University of Oklahoma, all to J.Z.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002780117/-/DCSupplemental.
Data Availability.
Genomic microarray data have been deposited in Gene Expression Omnibus (accession no. GSE98512).
References
- 1.Arneth A., et al. , Terrestrial biogeochemical feedbacks in the climate system. Nat. Geosci. 3, 525–532 (2010). [Google Scholar]
- 2.Oertel C., Matschullat J., Zurba K., Zimmermann F., Erasmi S., Greenhouse gas emissions from soils—A review. Geochemistry 76, 327–352 (2016). [Google Scholar]
- 3.Lee T. D., Barrott S. H., Reich P. B., Photosynthetic responses of 13 grassland species across 11 years of free-air CO2 enrichment is modest, consistent and independent of N supply. Glob. Change Biol. 17, 2893–2904 (2011). [Google Scholar]
- 4.Matamala R., Schlesinger William H., Effects of elevated atmospheric CO2 on fine root production and activity in an intact temperate forest ecosystem. Glob. Change Biol. 6, 967–979 (2001). [Google Scholar]
- 5.Carol A. E., Reich P. B., Trost J. J., Hobbie S. E., Elevated CO2 stimulates grassland soil respiration by increasing carbon inputs rather than by enhancing soil moisture. Glob. Change Biol. 17, 3546–3563 (2011). [Google Scholar]
- 6.Liu S., et al. , Climatic role of terrestrial ecosystem under elevated CO2 : A bottom-up greenhouse gases budget. Ecol. Lett. 21, 1108–1118 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Heath J., et al. , Rising atmospheric CO2 reduces sequestration of root-derived soil carbon. Science 309, 1711–1713 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Bradford M. A., et al. , Managing uncertainty in soil carbon feedbacks to climate change. Nat. Clim. Chang. 6, 751–758 (2016). [Google Scholar]
- 9.Li Y., et al. , Increasing importance of deposition of reduced nitrogen in the United States. Proc. Natl. Acad. Sci. U.S.A. 113, 5874–5879 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L., et al. , Different responses of soil respiration and its components to nitrogen addition among biomes: A meta-analysis. Glob. Change Biol. 20, 2332–2343 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Chen Z., et al. , Extreme rainfall and snowfall alter responses of soil respiration to nitrogen fertilization: A 3-year field experiment. Glob. Change Biol. 23, 3403–3417 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Janssens I. A., et al. , Reduction of forest soil respiration in response to nitrogen deposition. Nat. Geosci. 3, 315–322 (2010). [Google Scholar]
- 13.Olsson P., Linder S., Giesler R., Högberg P., Fertilization of boreal forest reduces both autotrophic and heterotrophic soil respiration. Glob. Change Biol. 11, 1745–1753 (2005). [Google Scholar]
- 14.Ward D., Kirkman K., Hagenah N., Tsvuura Z., Soil respiration declines with increasing nitrogen fertilization and is not related to productivity in long-term grassland experiments. Soil Biol. Biochem. 115, 415–422 (2017). [Google Scholar]
- 15.Sun Z., et al. , The effect of nitrogen addition on soil respiration from a nitrogen-limited forest soil. Agric. For. Meteorol. 197, 103–110 (2014). [Google Scholar]
- 16.Peng Q., et al. , Effects of nitrogen fertilization on soil respiration in temperate grassland in Inner Mongolia, China. Environ. Earth Sci. 62, 1163–1171 (2011). [Google Scholar]
- 17.Qi Y., et al. , Differential responses of short-term soil respiration dynamics to the experimental addition of nitrogen and water in the temperate semi-arid steppe of Inner Mongolia, China. J. Environ. Sci. (China) 26, 834–845 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Stocker T. F., Qin D., Plattner G.-K., Tignor M. M. B., IPCC, 2013: Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate (Cambridge University Press, Cambridge, UK, 2013), p. 1535. [Google Scholar]
- 19.Deng Q., et al. , Responses of soil respiration to elevated carbon dioxide and nitrogen addition in young subtropical forest ecosystems in China. Biogeosciences 7, 315–328 (2010). [Google Scholar]
- 20.Deng Q., et al. , Seasonal responses of soil respiration to elevated CO2 and N addition in young subtropical forest ecosystems in southern China. Ecol. Eng. 61, 65–73 (2013). [Google Scholar]
- 21.Craine Joseph M., Wedin David A., Reich Peter B., The response of soil CO2 flux to changes in atmospheric CO2, nitrogen supply and plant diversity. Glob. Change Biol. 7, 947–953 (2001). [Google Scholar]
- 22.Melillo J. M., et al. , Soil warming, carbon-nitrogen interactions, and forest carbon budgets. Proc. Natl. Acad. Sci. U.S.A. 108, 9508–9512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R., et al. , Soil C and N availability determine the priming effect: microbial N mining and stoichiometric decomposition theories. Glob. Change Biol. 20, 2356–2367 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Reich P. B., et al. , Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature 440, 922–925 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Fontaine S., et al. , Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 450, 277–280 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Adair E. C., Reich P. B., Hobbie S. E., Knops J. M., Interactive effects of time, CO2, N, and diversity on total belowground carbon allocation and ecosystem carbon storage in a grassland community. Ecosystems 12, 1037–1052 (2009). [Google Scholar]
- 27.Litton C. M., Raich J. W., Ryan M. G., Carbon allocation in forest ecosystems. Glob. Change Biol. 13, 2089–2109 (2007). [Google Scholar]
- 28.Cheng W., et al. , Synthesis and modeling perspectives of rhizosphere priming. New Phytol. 201, 31–44 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Dijkstra F. A., Carrillo Y., Pendall E., Morgan J. A., Rhizosphere priming: A nutrient perspective. Front. Microbiol. 4, 216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carreiro M., Sinsabaugh R., Repert D., Parkhurst D., Microbial enzyme shifts explain litter decay responses to simulated nitrogen deposition. Ecology 81, 2359–2365 (2000). [Google Scholar]
- 31.Wild B., et al. , Input of easily available organic C and N stimulates microbial decomposition of soil organic matter in arctic permafrost soil. Soil Biol. Biochem. 75, 143–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavicchioli R., et al. , Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowther T., et al. , The global soil community and its influence on biogeochemistry. Science 365, eaav0550 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., et al. , Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Chang. 2, 106–110 (2011). [Google Scholar]
- 35.Guo X., et al. , Gene-informed decomposition model predicts lower soil carbon loss due to persistent microbial adaptation to warming. Nat. Commun. 11, 4897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dijkstra F. A., Hobbie S. E., Reich P. B., Knops J. M. H., Divergent effects of elevated CO2, N fertilization, and plant diversity on soil C and N dynamics in a grassland field experiment. Plant Soil 272, 41–52 (2005). [Google Scholar]
- 37.Reich P. B., et al. , Do species and functional groups differ in acquisition and use of C, N and water under varying atmospheric CO2 and N availability regimes? A field test with 16 grassland species. New Phytol. 150, 435–448 (2001). [Google Scholar]
- 38.Zak D. R., Holmes W. E., Finzi A. C., Norby R. J., Schlesinger W. H., Soil nitrogen cycling under elevated CO2: A synthesis of forest face experiments. Ecol. Appl. 13, 1508–1514 (2003). [Google Scholar]
- 39.Melillo J. M., et al. , Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science 358, 101–105 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Melillo J. M., et al. , Soil warming and carbon-cycle feedbacks to the climate system. Science 298, 2173–2176 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Reich P. B., Hobbie S. E., Decade-long soil nitrogen constraint on the CO2 fertilization of plant biomass. Nat. Clim. Chang. 3, 278–282 (2013). [Google Scholar]
- 42.Zhou X., et al. , Concurrent and lagged impacts of an anomalously warm year on autotrophic and heterotrophic components of soil respiration: A deconvolution analysis. New Phytol. 187, 184–198 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Zhu B., Wang Y., Zheng X., Field measures of the contribution of root respiration to soil respiration in an alder and cypress mixed plantation by two methods: Trenching method and root biomass regression method. Eur. J. For. Res. 127, 285–291 (2008). [Google Scholar]
- 44.Kuzyakov Y., Larionova A. A., Root and rhizomicrobial respiration: A review of approaches to estimate respiration by autotrophic and heterotrophic organisms in soil. J. Plant Nutr. Soil Sci. 168, 503–520 (2005). [Google Scholar]
- 45.Atkin O. K., Bruhn D., Hurry V. M., Tjoelker M. G., The hot and the cold: Unravelling the variable response of plant respiration to temperature. Funct. Plant Biol. 32, 87–105 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Reich P. B., Walters M. B., Tjoelker M. G., Vanderklein D., Buschena C., Photosynthesis and respiration rates depend on leaf and root morphology and nitrogen concentration in nine boreal tree species differing in relative growth rate. Funct. Ecol. 12, 395–405 (1998). [Google Scholar]
- 47.Craine J. M., Wedin D. A., Chapin F. S., Reich P. B., Relationship between the structure of root systems and resource use for 11 North American grassland plants. Plant Ecol. 165, 85–100 (2003). [Google Scholar]
- 48.Carey J. C., et al. , Temperature response of soil respiration largely unaltered with experimental warming. Proc. Natl. Acad. Sci. U.S.A. 113, 13797–13802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu Q., et al. , GeoChip 4: A functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol. Ecol. Resour. 14, 914–928 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Xue K., et al. , Annual removal of aboveground plant biomass alters soil microbial responses to warming. mBio 7, e00976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hessen D. O., Ågren G. I., Anderson T. R., Elser J. J., de Ruiter P. C., Carbon sequestration in ecosystems: The role of stoichiometry. Ecology 85, 1179–1192 (2004). [Google Scholar]
- 52.Fontaine S., Mariotti A., Abbadie L., The priming effect of organic matter: A question of microbial competition? Soil Biol. Biochem. 35, 837–843 (2003). [Google Scholar]
- 53.Mamilov A. S., Dilly O. M., Soil microbial eco-physiology as affected by short-term variations in environmental conditions. Soil Biol. Biochem. 34, 1283–1290 (2002). [Google Scholar]
- 54.Locey K. J., et al. , Dormancy dampens the microbial distance-decay relationship. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue K., et al. , Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nat. Clim. Chang. 6, 595–600 (2016). [Google Scholar]
- 56.Davidson E. A., Samanta S., Caramori S. S., Savage K., The dual Arrhenius and Michaelis–Menten kinetics model for decomposition of soil organic matter at hourly to seasonal time scales. Glob. Change Biol. 18, 371–384 (2012). [Google Scholar]
- 57.Wang G., et al. , Soil moisture drives microbial controls on carbon decomposition in two subtropical forests. Soil Biol. Biochem. 130, 185–194 (2019). [Google Scholar]
- 58.Scurlock J., Hall D., The global carbon sink: A grassland perspective. Glob. Change Biol. 4, 229–233 (1998). [Google Scholar]
- 59.Reich P. B., et al. , Plant diversity enhances ecosystem responses to elevated CO2 and nitrogen deposition. Nature 410, 809–812 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Maier M., Schack-Kirchner H., Hildebrand E. E., Schindler D., Soil CO2 efflux vs. soil respiration: Implications for flux models. Agric. For. Meteorol. 151, 1723–1730 (2011). [Google Scholar]
- 61.Zhou J., Bruns M. A., Tiedje J. M., DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y., et al. , Responses of the functional structure of soil microbial community to livestock grazing in the Tibetan alpine grassland. Glob. Change Biol. 19, 637–648 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Weng E., Luo Y., Relative information contributions of model vs. data to short- and long-term forecasts of forest carbon dynamics. Ecol. Appl. 21, 1490–1505 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Xu R., Measuring explained variation in linear mixed effects models. Stat. Med. 22, 3527–3541 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Bland J. M., Altman D. G., Measurement error. BMJ 312, 1654 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moser E., Saxton A., Pezeshki S., Repeated measures analysis of variance: Application to tree research. Can. J. For. Res. 20, 524–535 (1990). [Google Scholar]
- 67.Alberti G., perm.t.test: R function for permutation-based t-test. https://rdrr.io/cran/GmAMisc/man/perm.t.test.html (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic microarray data have been deposited in Gene Expression Omnibus (accession no. GSE98512).