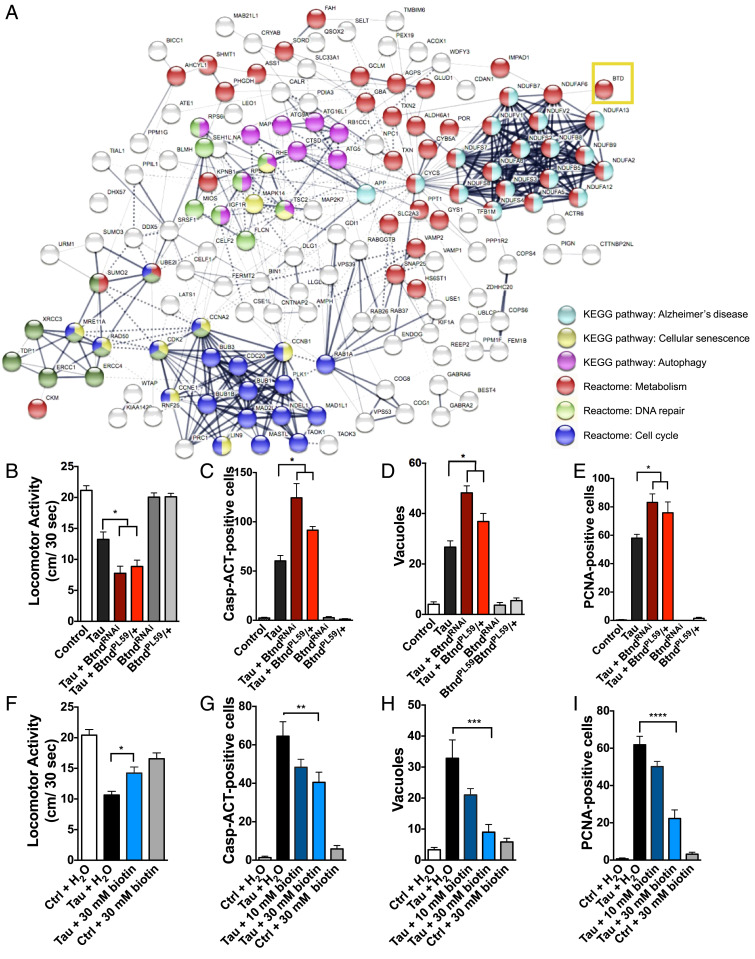
Fig. 1.
Pathway interactome of genetic hits from Drosophila RNAi screen of modifiers of tauR406W-mediated neurotoxicity and validation that biotin status modifies tau-mediated behavioral deficits and toxicity. (A) Predicted interactome from human orthologs of screen hits as shown by STRING analysis, including computationally predicted interactors from public datasets of direct physical and indirect functional interactions. Specific pathways enriched within the interactome are highlighted to the right. Validated screen hits were converted to human orthologs by DIOPT. Biotinidase gene is labeled as the human gene BTD (yellow box). (B) TauR406W-mediated locomotor deficits are enhanced by reduced biotinidase (BtndRNAi or BtndPL59/+) (ANOVA). (C–E) TauR406W-mediated neurotoxicity is enhanced by reduced biotinidase as shown by caspase activation, vacuole formation, and PCNA, an indicator of cell-cycle reentry (ANOVA). (F) TauR406W-mediated locomotor deficits are rescued by biotin feeding. (G–I) TauR406W-mediated neurotoxicity is rescued by biotin feeding as shown by caspase activation, vacuole formation, and PCNA. Biotin was mixed into Drosophila instant food and replaced every third day. All flies were 10 d old at time of experimentation. n = 6 per genotype (two-way ANOVA + Tukey’s post test). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Control genotype B, D, E, F, H, and I: elav-GAL4/+. Control genotype C, G: elav-GAL4/+; UAS-CD8-PARP-Venus/+.