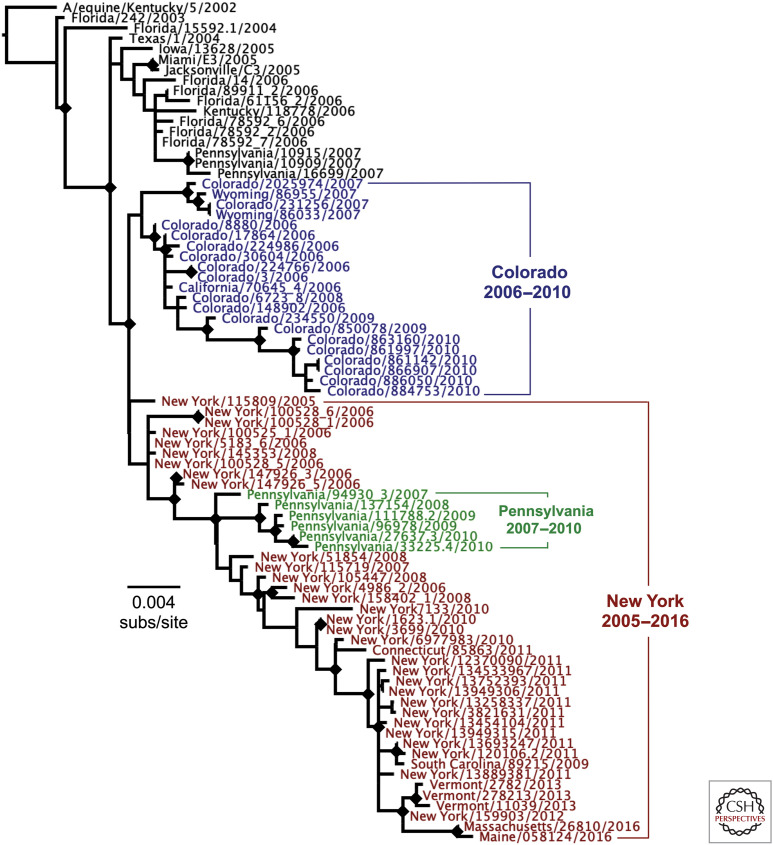
Figure 2.
Evolutionary relationships among the hemagglutinins (HAs) of the H3N8 canine influenza virus (CIV) since the emergence of that virus in dogs. All available HA sequences were used in the analysis and were outgroup-rooted on a 2002 H3N8 equine influenza virus HA sequence (strain A/equine/Kentucky/5/2002). Phylogenetic relationships were determined using the maximum likelihood (ML) method available in PhyML (Guindon and Gascuel 2003), using a general time-reversible (GTR) substitution model, gamma-distrusted (Γ) rate variation among sites, and bootstrap resampling (100 replicates). Diamonds at nodes indicate bootstrap support in ≥70 out of 100 replicates. Regional lineages were seen to develop in Colorado (blue) and in New York (red) and nearby states in the United States, as well as a sublineage that arose around Philadelphia (green) between 2007 and 2010.