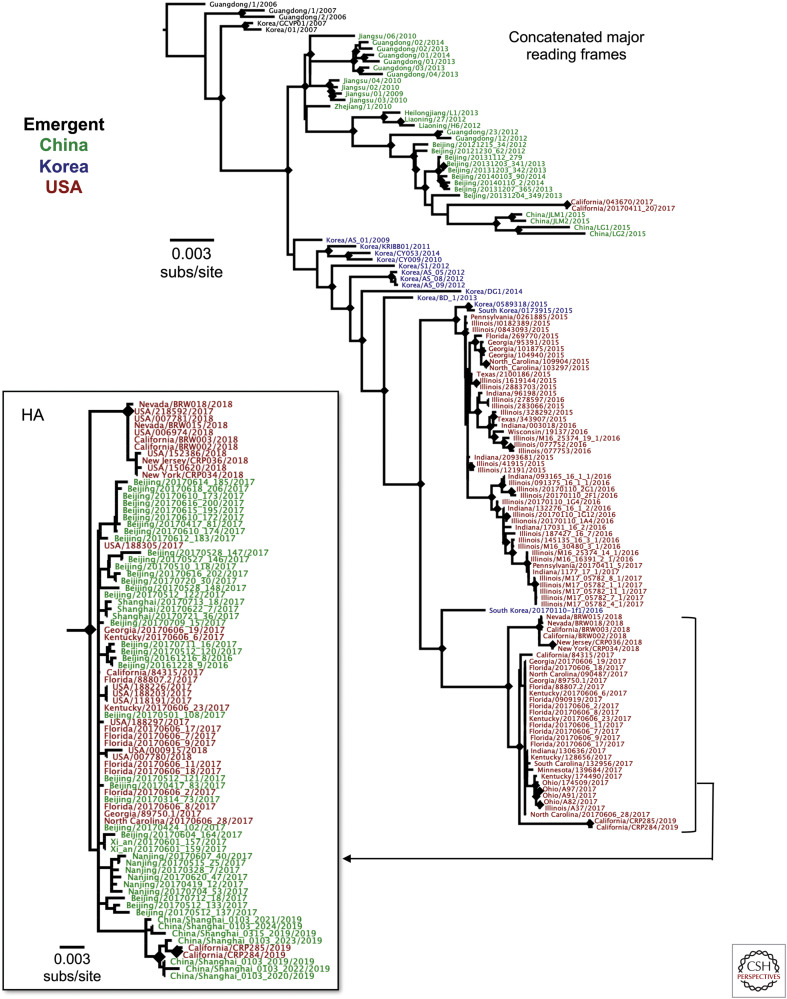
Figure 3.
Evolutionary relationships among H3N2 canine influenza virus (CIV) in China, Korea, and the United States, showing how the viruses spread among the different regions. An analysis of available full genome sequences, in which the eight genome segment open reading frames are concatenated. The tree is outgroup-rooted on the earliest available H3N2 CIV sequence in the database (A/canine/Guangdong/1/2006). Just eight of 149 genome sequences contained reassortant viruses, as determined by RDP4 analysis (Martin et al. 2015), and were removed before analysis. Phylogenetic relationships were determined using the maximum likelihood (ML) method available in PhyML (Guindon and Gascuel 2003), using a general time-reversible (GTR) substitution model, gamma-distrusted (Γ) rate variation among sites, and bootstrap resampling (100 replicates). Diamonds at nodes indicate bootstrap support in ≥70 of 100 replicates. Colors indicate countries of origin from Korea (blue), China (green), or the United States (red). An analysis of the hemagglutinin (HA) segment sequences (inset) available for 2017–2019 isolates of H3N2 CIV show the close relationships between those viruses in the United States and China.