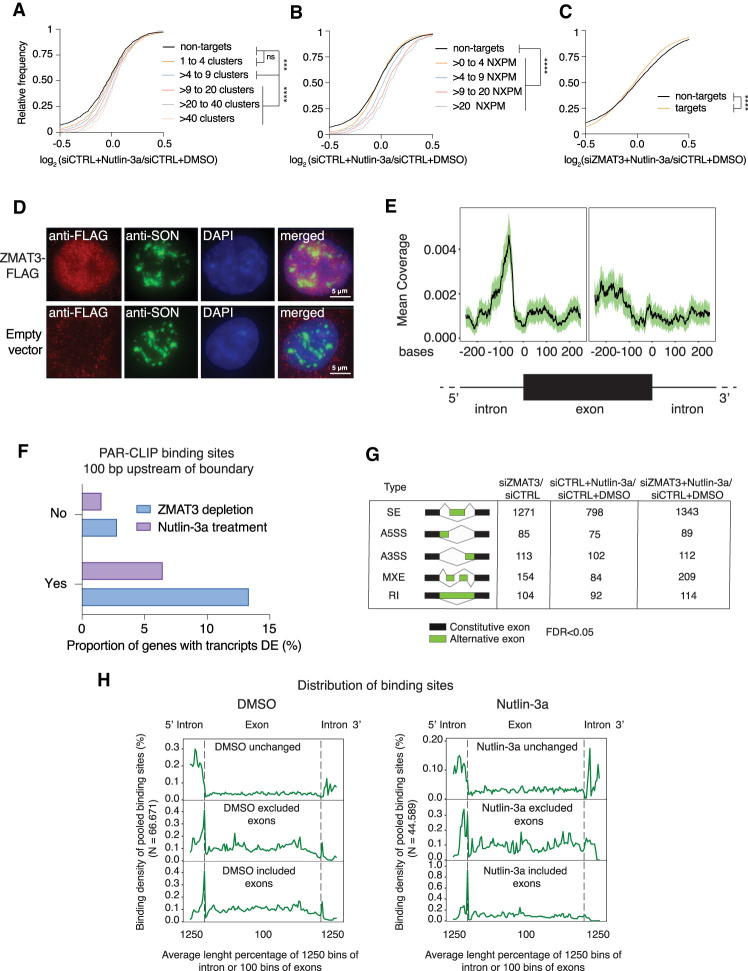
Figure 2.
ZMAT3 predominantly binds to regions close to 3′ splice sites and its knockdown induces widespread changes in splicing patterns. (A,B) mRNA expression changes upon ZMAT3 induction in HCT116 cells determined by RNA-seq in biological triplicates. The empirical cumulative distribution function of ZMAT3 PAR-CLIP targets (from HCT116 cells after Nutlin-3a treatment) binned by number of binding sites (A) or number of NXPM (cross-linked reads per million normalized to target gene expression levels) (B) compared with nontargets (FPKM > 5, black line). P-values were determined by Kolmogorov-Smirnov test (***) P < 0.001, (****) P < 0.0001, (ns) nonsignificant. (C) Same as A after knockdown of ZMAT3 and only separating ZMAT3 target genes (orange line) from nontargets (black line). (D) Fluorescent microscopy images of immunostaining experiments with HCT116-ZMAT3-FLAG (top images) and HCT116-FLAG cells stained with antibodies against FLAG (red) or the nuclear speckle marker protein SON (green). DNA is counterstained with DAPI (blue). (E) Plot of cross-linked sequence read coverage distributed along the intron-exon boundary from a representative sample of ZMAT3 PAR-CLIP from DMSO treated HCT116 cells. (F) Percentage of ZMAT3 target mRNAs with significantly changed abundance (P<0.05, Sleuth) in their transcripts that contained (Yes) or did not contain (No) ZMAT3-binding sites within 100 bp of the 3′ splice site. Note that the number of gene expression changes was significantly higher in HCT116 cells after ZMAT3 knockdown (kd) compared with ZMAT3 induction by Nutlin-3a. (DE) Differentially expressed. (G) Table summarizing alternative splicing events (FDR < 0.05, reads ≥ 5 and ΔPSI ≥ 10%) upon ZMAT3 kd, Nutlin-3a treatment, or ZMAT3 kd prior to Nutlin-3a treatment, compared with control HCT116 cells. SE, skipped exons; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site; MXE, mutually exclusive exon; RI, retained introns. (H) Metagene plot of cross-linked sequence reads at the intron-exon boundary from pooled ZMAT3 PAR-CLIPs in DMSO- or Nutlin-3a-treated cells. The metagene plots were separated according to whether the exons were excluded, included, or unchanged upon ZMAT3 kd.