In this study, Zhu et al. sought to understand the signaling pathways and epigenetic control of nuclear architecture during developmental progression of immune cells. They found that in activated neutrophils calcium influx rapidly recruited the cohesin-loading factor NIPBL to thousands of active enhancers and promoters to dictate widespread changes in compartment segregation.
Keywords: chromatin remodelers, cohesin, enhancers, NIPBL, neutrophils, P300 and CBP, promoters
Abstract
During developmental progression the genomes of immune cells undergo large-scale changes in chromatin folding. However, insights into signaling pathways and epigenetic control of nuclear architecture remain rudimentary. Here, we found that in activated neutrophils calcium influx rapidly recruited the cohesin-loading factor NIPBL to thousands of active enhancers and promoters to dictate widespread changes in compartment segregation. NIPBL recruitment to enhancers and promoters occurred with distinct kinetics. The induction of NIPBL-binding was coordinate with increased P300, BRG1 and RNA polymerase II occupancy. NIPBL-bound enhancers were associated with NFAT, PU.1, and CEBP cis elements, whereas NIPBL-bound promoters were enriched for GC-rich DNA sequences. Using an acute degradation system, we found that the histone acetyltransferases P300 and CBP maintained H3K27ac abundance and facilitated NIPBL occupancy at enhancers and that active transcriptional elongation is essential to maintain H3K27ac abundance. Chromatin remodelers, containing either of the mutually exclusive BRG1 and BRM ATPases, promoted NIPBL recruitment at active enhancers. Conversely, at active promoters, depletion of BRG1 and BRM showed minimal effect on NIPBL occupancy. Finally, we found that calcium signaling in both primary innate and adaptive immune cells swiftly induced NIPBL occupancy. Collectively, these data reveal how transcriptional regulators, histone acetyltransferases, chromatin remodelers, and transcription elongation promote NIPBL occupancy at active enhancers while the induction of NIPLB occupancy at promoters is primarily associated with GC-rich DNA sequences.
It is now well established that the mammalian genome is highly structured. Chromatin is organized as loop domains or topologically associating domains, that fold into either the transcriptionally active (A) compartment or repressive (B) compartment (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Gibcus and Dekker 2013; Rao et al. 2014; Rowley and Corces 2018). Loop domains are established by cohesin-mediated extrusion of chromatin. Cohesin is loaded onto chromatin primarily by NIPBL and MAU2 (Ciosk et al. 2000; Nasmyth and Haering 2009; Murayama and Uhlman 2014; Petela et al. 2018; Parenti et al. 2020). Once loaded, cohesin progressively extrudes chromatin loops until a pair of convergent CTCF sites is reached (Kueng et al. 2006; Rao et al. 2014; Sanborn et al. 2015; Fudenberg et al. 2016; Haarhuis et al. 2017; Wutz et al. 2017; Vian et al. 2018). Release of cohesin from chromatin is orchestrated by WAPL (Kueng et al. 2006; Haarhuis et al. 2017; Wutz et al. 2017). NIPBL and cohesin mediated formation of loop domains suppress compartmental segregation allowing for fine-scale control over local chromatin interactions and regulation of gene expression (Haarhuis and Rowland 2017; Rao et al. 2017; Schwarzer et al. 2017; Nuebler et al. 2018).
Neutrophils are innate antimicrobial effector cells that rapidly respond to a diverse range of extracellular stimuli. Upon microbial encounter, neutrophils activate inflammatory gene programs, engulf bacteria through phagocytosis, discharge microbicidal factors into the extracellular space, and release an extracellular network of chromatin fibers to trap invading pathogens (Brinkmann et al. 2004; Tecchio et al. 2014; Ley et al. 2018). The activation of neutrophils involves both calcium-independent and calcium-dependent mechanisms (Kenny et al. 2017). Intracellular calcium levels are tightly controlled and elevated upon stimulation (Hogan et al. 2010). A prominent target of calcium is nuclear factor of activated T cells (NFAT) that comprises a family of transcription factors (TFs) important for regulating T and B cell activation as well as innate immune responses (Fric et al. 2012; Vaeth et al. 2018). Inactive NFAT proteins are generally excluded from the nucleus through the phosphorylation of specific serine residues. Calcium influx activates the phosphatase calcineurin, which removes the regulatory phosphate groups and allows the translocation of NFAT into the nucleus (Hogan et al. 2003, 2010).
In recent studies we demonstrated that upon activation human neutrophils rapidly sequester cohesin at H3K27ac-marked enhancers (Denholtz et al. 2020). Here we examined in neutrophils whether and how calcium signaling modulates nuclear architecture. We found that calcium signaling triggered rapid changes in compartment segregation. Calcium-induced alterations in compartmentalization were closely associated with calcium-induced NIPBL occupancy. Calcium-induced recruitment of NIPBL involved a wide spectrum of TFs, including NFAT and PU.1. The induction of NIPBL-binding was coordinate with increased co-occupancy of the histone acetyltransferase (HAT) P300, the chromatin remodeler BRG1, and RNA polymerase II (RNAPII). To delineate the pathway that underpins calcium-induced NIPBL recruitment we generated an armamentarium of cell lines that carried inducible degrons. We found that acute degradation of P300- and CREB-binding protein (CBP) or SPT5 reduced H3K27ac abundance and decreased genome-wide NIPBL occupancy. Acute degradation of both BRG1 and BRM eliminated calcium-induced NIPBL occupancy at active enhancers but not at promoters. Finally, we found that calcium signaling orchestrated lineage-specific NIPBL occupancy in primary innate and adaptive immune cells. Collectively, these data reveal that active transcriptional elongation maintains H3K27ac abundance and show in mechanistic terms how calcium influx dictates NIPBL occupancy at active enhancers and promoters to orchestrate changes in nuclear architecture.
Results
Calcium influx triggers a weakening of compartment segregation
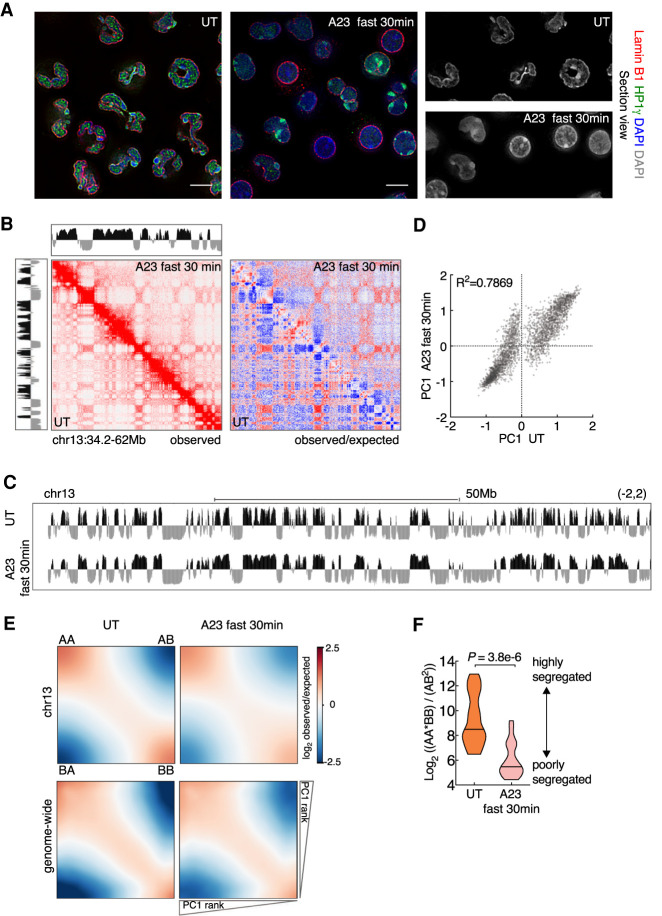
In previous studies we demonstrated that the genomes of mature neutrophils are characterized by a highly segregated compartment structure (Zhu et al. 2017; Denholtz et al. 2020). We also showed that exposure of human neutrophils to microbial agents readily induced recruitment of cohesin to H3K27ac marked enhancers (Denholtz et al. 2020). As a model system to determine how signals emanating from the cell surface instruct alterations in nuclear morphology we used differentiated neutrophils derived from an immortalized neutrophil progenitor cell line, ECOMG (Zhu et al. 2017). To study the impact of activation on neutrophil genome topology, ECOMG-derived neutrophils were cultured in the presence of the calcium ionophore A23187 for different durations and using different concentrations. Activated neutrophils were examined for changes in nuclear morphology using DAPI, Lamin B1, and HP1γ staining. We found that when treated with 5 µM A23187 for a duration of 4 h, neutrophils displayed a loss of lobular nuclear structures (data not shown). Treatment with higher concentrations of A23187 (20 µM) for 30 min resulted in a robust and homogeneous loss of lobular nuclear structures and spreading of condensed chromatin from the nuclear periphery across the entire nucleoplasm (Fig. 1A).
Figure 1.
Calcium entry triggers a weakening of compartmentalization. (A) Immunofluorescence images comparing nuclear morphology of untreated (UT) and A23187-activated (A23187 fast 30 min) neutrophils. Neutrophils were activated using fast activation condition (20 µM A23187) for 30 min. Nuclear lamina was stained using anti-LaminB1. Anti-HP1γ stained HP1γ that binds both euchromatin and heterochromatin. Counterstain with DAPI highlighted heterochromatin. Representative image sections are presented as digitally magnified images. Original magnification, ×100. Scale bars, 5 µm. (B) Hi-C contact matrices for chromosome 13 (34.2–62 Mb) in untreated and activated neutrophils. The intensity of each pixel represents the normalized number of contacts between a pair of loci. (Left) Observed contact maps were balanced normalized at 50-kb resolution with a color range of 0–20. (Right) Observed/expected contact maps were balanced normalized and distance normalized at 50-kb resolution with a color range of −4 to 4 (log2 ratio). Corresponding compartment profiles (PC1 tracks) at 25-kb resolution, indicating genomic regions positioned in the A (black) and B (gray) compartments are shown above and to the left of the matrix for activated and untreated neutrophils, respectively. (C) Genome browser tracks for PC1 spanning chromosome 13 in untreated and activated neutrophils. (D) Scatter plot for PC1 values of untreated versus activated neutrophils, defined at PC1 domain (PD) level is shown. Signals away from the diagonal indicate PDs that switched compartments. Pearson coefficients are indicated. (E) Saddle plots for compartmentalization strength in untreated (left panels) and A23187-activated (right panels) neutrophils. Average distance-normalized contact frequencies between cis pairs of 50-kb bins arranged by descending PC1 values. The top shows saddle plots for chr13. The bottom shows aggregated map constructed by averaging all chromosomes. (F) Violin plots comparing distributions of log2 ratios for average genomic interaction frequencies involving different compartments (AB) compared with those involving the same compartments (AA and BB). The median values are shown as central black lines. P-value, two-tailed Wilcoxon rank-sum test.
To examine how calcium signaling modulates genome topology we used in situ Hi-C. More than 180 million valid contacts were generated for each condition (Supplemental Table S1). We found that contact frequencies as a function of genomic distance scaled similarly for resting and activated neutrophils (Supplemental Fig. S1A). However, we found that contact matrices revealed substantial differences in long-range genomic interactions when comparing naïve versus activated neutrophils (Fig. 1B). Resting neutrophils were associated with a plaid-pattern involving long-range cis contacts in Hi-C contact matrices that were weakened in activated neutrophils (Fig. 1B). We next examined A and B compartment segregation using principal component analysis of normalized Hi-C matrices. Whereas resting neutrophil genomes were associated with highly segregated compartments, activated neutrophils revealed at the boundaries of small compartment domains a striking loss of compartmentalization, with small B regions converging into A regions and small A regions converging into B regions (Fig. 1C,D; Supplemental Fig. S1B). The loss of finely structured compartments resulted in a decrease in the numbers of principal domains (PDs) in A and B compartments (Supplemental Fig. S1C). PDs that switched compartments upon neutrophil activation were on average of smaller size than those that did not undergo changes in compartmentalization (Supplemental Fig. S1D,E; Supplemental Table S2). Upon calcium-induced neutrophil activation, PDs that switched compartments showed intermediate PC1 values (Supplemental Fig. S1F; Supplemental Table S2). The number of genomic regions that converted from the B to the A compartment was significantly higher than those that repositioned from the A to B compartment. We quantified the weakening of compartment segregation by calculating compartment strength. We found that calcium signaling depleted for AA and BB interactions whereas AB interactions were enriched (Fig. 1E). We validated this analysis by plotting the ratios for average interaction frequencies involving the same compartment (AA and BB) versus those involving different compartments (AB). This analysis validated the notion that calcium signaling diminishes segregation of A and B compartments (Fig. 1F).
Previously we found that during neutrophil differentiation A and B PDs that span large genomic distances fragmented into smaller domains (Zhu et al. 2017). Here we found that calcium entry in neutrophils induced a reversed pattern: highly segmented compartments that merged into singular compartments resembling that of neutrophil progenitors. To explore whether and how changes in compartmentalization and epigenetic marking are related, we examined resting and activated neutrophils for the deposition of H3K27me3 and H3K36me3. We found that genomic regions that switched from A to B compartments were initially associated with actively transcribed regions (H3K27me3lowH3K36me3hi), while genomic regions that switched from B to A compartments were associated with transcriptionally silent genomic regions (H3K27me3hiH3K36me3low) (Supplemental Fig. S1G–J). Finally, we compared the PC1 values of PDs that upon activation switched compartments to that of other cell types. We found that PDs that upon neutrophil activation repositioned from A to B compartments were predominantly located in the B compartment in other immune cells (Supplemental Fig. S1K). Conversely, PDs that switched from B to A compartments were primarily localized in the A compartments in other immune cells (Supplemental Fig. S1K). In sum, these data indicate that in neutrophils calcium entry induced a weakening of compartment segregation to establish a topology akin to that of other immune cells.
Calcium signaling elevates NIPBL and SMC1 occupancy at active enhancers and promoters
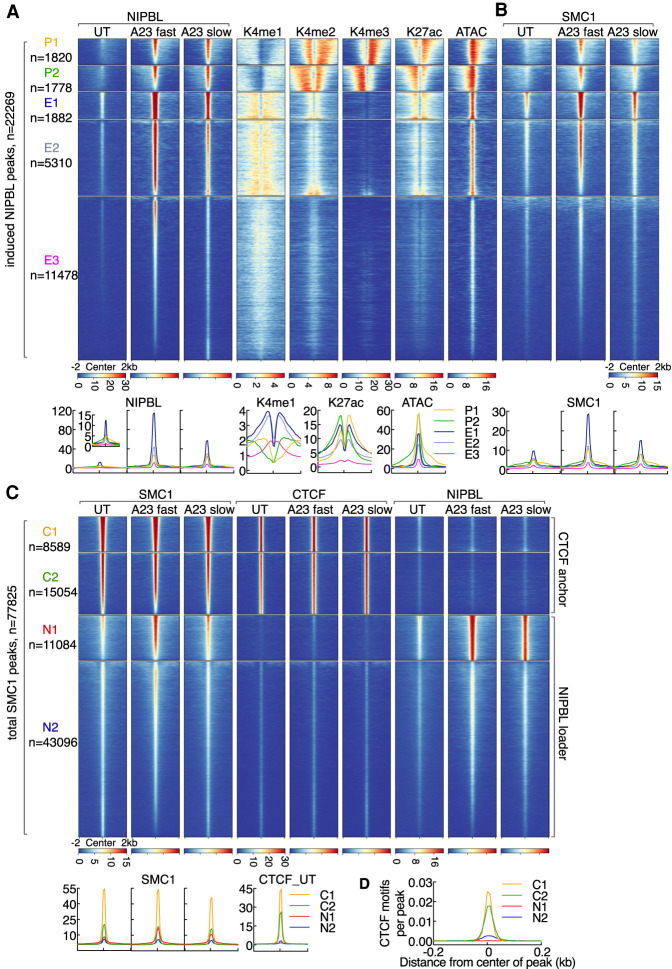
Recent studies demonstrated that chromatin organization emerges from competition of nonequilibrium active loop extrusion and epigenetically defined compartment structure (Haarhuis and Rowland 2017; Rao et al. 2017; Schwarzer et al. 2017; Nuebler et al. 2018). To explore the possibility that calcium-induced loop extrusion suppressed compartment segregation we examined for NIPBL and SMC1 occupancy. We found that upon calcium entry, genome-wide NIPBL occupancy greatly increased (Supplemental Fig. S2A,B). NIPBL-bound sites in resting and activated neutrophils, using both fast and slow activating conditions, partially overlapped, indicating that calcium entry elevates NIPBL occupancy at pre-existing as well as de novo sites (Supplemental Fig. S2A). Among the 55,075 NIPBL-binding sites that were identified and merged from the aforementioned conditions, NIPBL occupancy at 22,269 sites was significantly elevated using either fast or slow activation conditions (FC > 4, P < 0.0001).
To investigate how NIPBL occupancy is distributed across the promoter and enhancer landscape, resting and activated neutrophils were examined for the deposition of epigenetic modifications that mark promoters (H3K4me3), poised enhancers (H3K4me1) and active promoters and enhancers (H3K27ac). Additionally, we examined resting and activated neutrophils for nucleosome density using ATAC-seq. K-means clustering revealed that calcium-induced NIPBL-bound sites were distributed as groups across the promoter (clusters P1 and P2) and enhancer landscapes (clusters E1, E2, and E3) (Fig. 2A; Supplemental Fig. S2B–D; Supplemental Table S3). NIPBL-bound promoters segregated into two clusters, P1 and P2, reflecting differences in transcriptional direction, while NIPBL-bound enhancers segregated into three clusters reflecting differences in the deposition of H3K27ac across active enhancers (E1 and E2) versus poised enhancers (E3) (Fig. 2A). Prior to activation, enhancers and promoters exhibited relatively low levels of NIPBL occupancy (Fig. 2A). Upon calcium entry NIPBL-binding strength was elevated at both active enhancers and promoters but to a much greater degree at enhancers (Fig. 2A). We found that induced NIPBL levels at enhancers were proportional to pre-existing levels of NIPBL (E1 > E2 > E3) and proportional to pre-existing H3K27ac abundance and chromatin accessibility (Fig. 2A). As expected, calcium-induced NIPBL occupancy was coordinate with increased SMC1 occupancy (Fig. 2B).
Figure 2.
Calcium signaling rapidly elevates NIPBL occupancy at active enhancers and promoters. (A) Heat map of K-means clustering analysis for NIPBL and epigenetic marks as revealed by ChIP-seq and ATAC-seq signals for ±2-kb windows centered on induced NIPBL-binding sites (n = 22,269, FC > 4, K = 5). Each cluster was manually classified as promoters (P1 and P2) or enhancers (E1, E2, and E3) according to epigenetic features. Numbers of NIPBL-binding sites for each cluster are denoted at the left. The five clusters were saved as a reference bed file to compute matrices for the following analysis. Heat maps were generated by sorting maximum signal intensities associated with NIPBL occupancy for neutrophils cultured in the presence of A23187 (fast activation conditions) and calculated across the entire 4-kb interval around peak summits. Color scale intensities represent normalized reads scores (reads per 10 million uniquely mapped reads per base pair). Note that the blue-yellow-red color scheme was used for ChIP-seq, GRO-seq, and ATAC-seq heat maps throughout the manuscript except for the heat map shown in Figure 5G. Corresponding average enrichment plots with mean signal intensities (per base pair per peak) are shown below. Neutrophils were activated either in fast activation condition (20 µM A23187 for 15 min), or in slow activation condition (5 µM A23187 for 4 h). These two activation conditions were used throughout the manuscript unless otherwise mentioned. (B) Heat map of SMC1 ChIP-seq signals plotted for five clusters defined in A. Heat map was sorted by the maximum signal intensity of SMC1 occupancy using A23187 fast activation conditions. Corresponding average enrichment plots are shown below the heat maps. (C) Heat map of K-means clustering analysis for SMC1, CTCF, and NIPBL ChIP-seq signals across a region of ±2 kb centered on total SMC1 peaks (n = 77,825, K = 4). Each cluster was manually classified as anchors or loaders according to its characteristic features. Heat map was sorted by the maximum signal intensity associated with SMC1 occupancy in A23187 fast activation conditions. Corresponding average enrichment plots are shown below the heat maps. (D) Average enrichment plots for mean motif densities (motifs per base pair per peak) associated with CTCF motifs, centered on CTCF-binding sites (±200 bp) are shown for clusters C1 and C2.
Consistent with previous studies, SMC1 bound sites were associated either with CTCF or NIPBL occupancy (Kagey et al. 2010; Vian et al. 2018). Using four-cluster K-means clustering analysis we identified two distinct groups of CTCF (C1 and C2) and NIPBL (N1 and N2) bound sites (Fig. 2C). Upon calcium influx, SMC1 occupancy at CTCF-bound sites essentially remained the same (Fig. 2C). In contrast, calcium-induced SMC1 occupancy was significantly elevated at NIPBL-bound sites (P = 1.82 × 10−12) (Fig. 2C). Finally, SMC1 levels at CTCF/SMC1-bound sites positively correlated with CTCF motif densities (C1 > C2) (Fig. 2D). SMC1 levels at NIPBL/SMC1 bound sites positively correlated with NIPBL-binding strength (N1 > N2) coordinate with enhancer and promoter activity (Fig. 2C). Taken together, these data indicate that upon calcium entry NIPBL and SMC1 are primarily sequestered at H3K27ac-marked promoters and H3K27ac-marked enhancers but not at H3K4me1-marked poised enhancers.
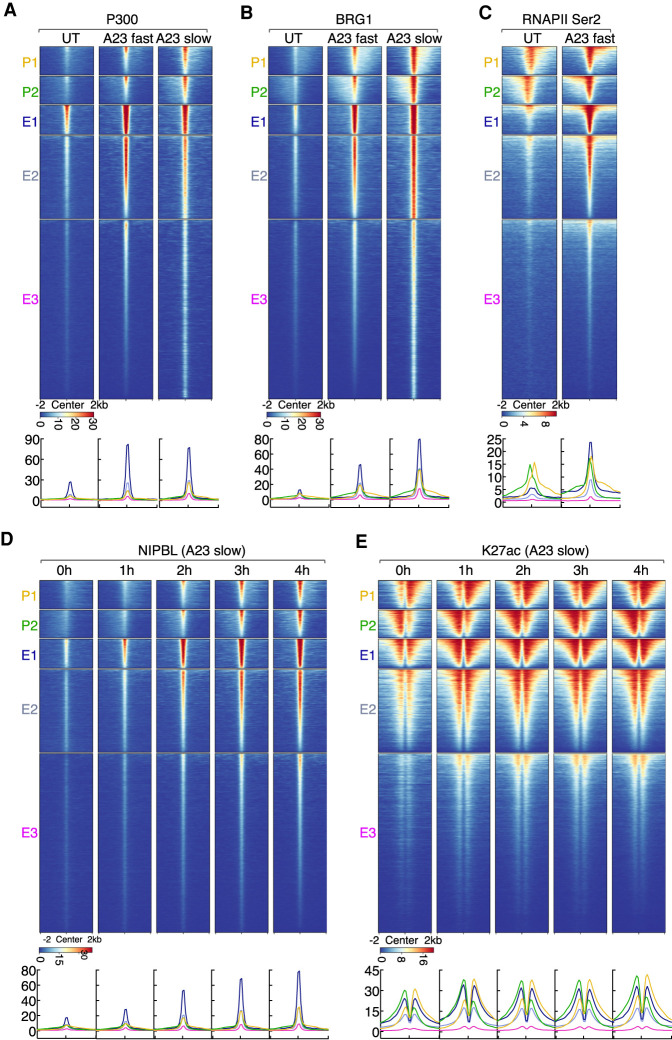
Calcium signaling instructs with distinct kinetics P300, BRG1, RNAPII, and NIPBL occupancy at enhancers versus promoters
To examine whether and how upon calcium influx NIPBL coordinately binds with P300, BRG1, and RNAPII to promoter and enhancer regions we used ChIP-seq. We found that prior to activation P300, BRG, RNAPII, and NIPBL showed low occupancy (Fig. 3A–D). Upon calcium influx, the induction of NIPBL occupancy was coordinate with P300, BRG1, and RNAPII binding (Fig. 3A–D). To determine the kinetics associated with NIPBL, BRG1, P300, and RNAPII Ser2 recruitment, we used slow activation conditions. We observed a gradual and sequential induction of NIPBL, BRG1, P300, and RNAPII Ser2 occupancy at active enhancers and promoters (Fig. 3D; Supplemental Fig. S3A,B). We found that NIPBL, BRG1, and P300 accumulated with faster kinetics at enhancers than at promoters (Fig. 3D; Supplemental Fig. S3A,B). At active enhancers, calcium influx activated a gradual recruitment of NIPBL that was initiated within 1 h to reach maximum levels 3–4 h after activation (Fig. 3D). Conversely at promoters, calcium-induced NIPBL recruitment did not occur until 3–4 h after stimulation (Fig. 3D). P300 and BRG1 occupancy mirrored the kinetics of NIPBL binding (Supplemental Fig. S3A,B). The kinetics of H3K27ac abundance at enhancers and promoters reached maximum levels 1 h after activation, which was maintained for the duration of calcium influx (Fig. 3E). RNAPII (RNAPII Ser2) rapidly associated with both active promoters and enhancers (Supplemental Fig. S3C). RNAPII Ser2 accumulation at active enhancers was particularly prominent using fast activation conditions (15 min) when compared with slow activation conditions (4 h), plausibly imposed by time-limited transcriptional elongation (Fig. 3C; Supplemental Fig. S3C). Taken together, these data indicate that calcium entry recruits NIPBL, P300, BRG1, and RNAPII Ser2 at active enhancers and promoters but with different kinetics.
Figure 3.
Calcium signaling rapidly elevates with distinctive kinetics P300, BRG1, and RNAPII co-occupancy at NIPBL-bound enhancers and promoters. (A) Heat map displaying P300 ChIP-seq signals plotted for five clusters of genomic regions defined in Figure 2A. Heat map was sorted by the maximum signal intensity of P300 in A23187 fast activation conditions. (B) Heat map displaying BRG1 ChIP-seq signals plotted for five clusters of genomic regions defined in Figure 2A is shown. Heat map was sorted by the maximum signal intensity of BRG1 in A23187 fast activation conditions. (C) Heat map displaying RNAPIISer2 ChIP-seq signals plotted for five clusters of genomic regions defined in Figure 2A. Heat map was sorted by the maximum signal intensity of RNAPII in A23187 fast activation conditions. (D) Kinetics of NIPBL recruitment is shown. Heat map of NIPBL ChIP-seq signals was plotted for five clusters of genomic regions defined in Figure 2A. Heat map was generated by centering on the maximum signal intensity of NIPBL occupancy in A23187 slow activation conditions (4 h). (E) Kinetics of H3K27ac distribution is shown. Heat map of H3K27ac ChIP-seq signals was plotted for five clusters of genomic regions defined in Figure 2A. Heat map was sorted by centering on the maximum signal intensity of H3K27ac in A23187 slow activation conditions (4 h). (A–E) Below the heat maps are indicated the corresponding average enrichment plots.
Upon calcium entry NIPBL is sequestered at active promoters and translocated coordinately with elongating RNA polymerase
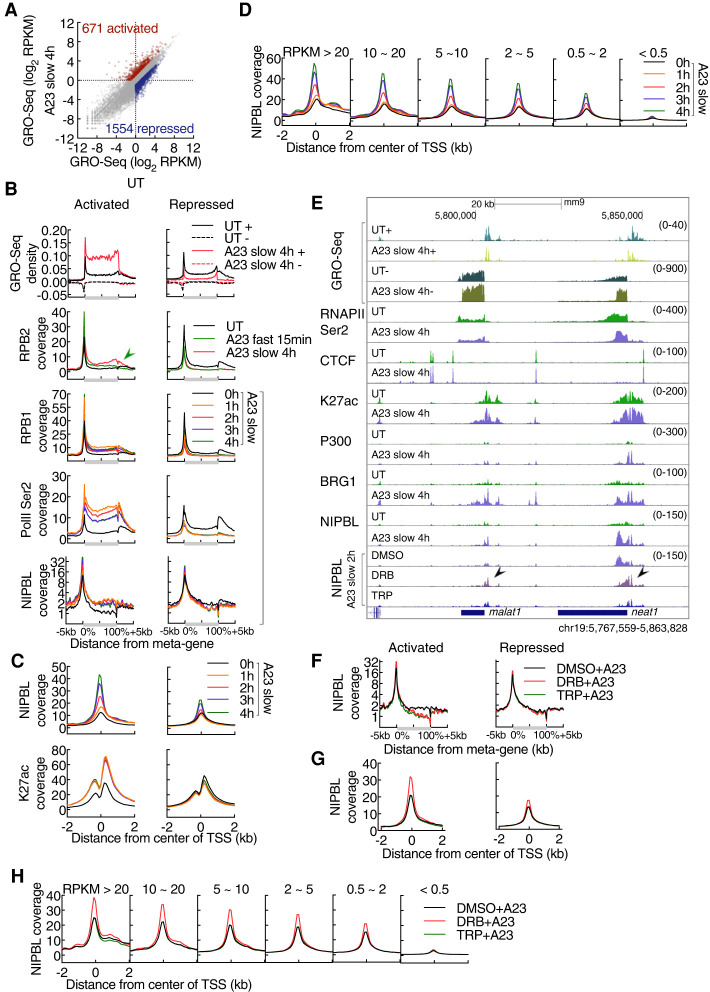
To determine whether and how nascent transcript abundance relates to NIPBL recruitment at promoters, neutrophils were activated for 4 h and analyzed using GRO-seq. We found that calcium signaling elevated the abundance of 671 transcripts, while 1554 transcripts showed decreased abundance (Fig. 4A; Supplemental Table S4). Prominent pathways that were affected included the NF-κB, inflammatory and cytokine mediated signaling cascades as well as TFs involved in the immediate stress response, including nr4a1-3, fos, and fosb (Supplemental Table S5). To determine how calcium entry modulates transcript abundance during the course of 1–4 h after calcium influx we performed RPB1 and RNAPIISer2 ChIP-seq. As expected, RBP1 and RNAPII Ser2 abundance differed when comparing naïve versus activated neutrophils (Supplemental Fig. S4A). For a duration of 1–4 h after activation, RBP1 and RNAPII Ser2 abundance remained equivalent (Supplemental Fig. S4A). At activated genes calcium entry prompted productive RNAPII elongation along gene bodies while RNAPII and RNAPII Ser2 were depleted from gene-bodies and promoters at repressed genes (Supplemental Fig. S4B). RBP1 and RNAPII Ser2 abundance reached maximum levels already 1 h after activation (Supplemental Fig. S4B,C). Meta-gene analysis revealed tightly correlated RPB1, RNAPII Ser2, and NIPBL occupancy (Fig. 4B). Upon activation, the deposition of H3K27ac at promoters rapidly increased to reach maximum levels at 1 h, while NIPBL occupancy steadily increased during the course of activation (Fig. 4C). NIPBL occupancy at promoters and gene bodies also correlated well with global mRNA abundance (Fig. 4D). To determine whether and how transcriptional initiation and elongation modulates calcium-induced NIPBL-occupancy, we cultured neutrophils in the presence of Triptolide (TRP) and 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside (DRB). Specifically, neutrophils were cultured for 2 h in the presence of inhibitors, followed by another 2 h in the presence of A23187. We found that in the presence of inhibitors calcium-induced NIPBL occupancy significantly accumulated at TSSs but was depleted across gene bodies (Fig. 4E–G; Zhou et al. 2012).
Figure 4.
Transcriptional elongation modulates NIPBL occupancy at promoters. (A) Scatter plot for GRO-seq read density of untreated versus activated neutrophils. Color-codes indicate a decrease (blue) or increase (red) in transcript abundance in activated versus naïve neutrophils (FC > 2, RPKM > 0.5). Numbers of genes are indicated in parenthesis. (B) Meta-gene profile of GRO-seq (tags per base pair per peak) and ChIP-seq signals (tags per base pair per peak) across genic regions in untreated and activated neutrophils, gated on differentially expressed genes as defined in A. Gene lengths (from TSS to TTS) were rescaled to the same pseudo-length as shown on the X-axis. Sense (+) and antisense (−) transcripts are plotted above and below the X-axis, respectively. Note that the green arrow shows an atypical distribution pattern for RNAPII at activated genes in response to fast activation. (C) Average enrichment plots for NIPBL and H3K27ac ChIP-seq signals in untreated and activated neutrophils, plotted as a function of genomic distance from the center of TSSs (±2 kb), gated on differentially expressed genes as defined in A. (D) Average enrichment plots for NIPBL ChIP-seq signals in untreated and activated neutrophils, plotted as a function of genomic distance from the center of TSSs (±2 kb). Genes are grouped according to expression levels in activated neutrophils defined by GRO-seq reads. (E) Genome browser tracks of GRO-seq and ChIP-seq signals in untreated and activated neutrophils, pretreated with or without transcription inhibitors for 2 h, for representative loci named malat1 and neat1 are shown. DRB refers to 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside. TRP refers to triptolide (TRP). Black arrowheads indicate NIPBL bound sites affected by DRB. (F) Meta-gene profile associated with NIPBL ChIP-seq reads across genic regions pretreated with or without transcription inhibitors for 2 h, gated on differentially expressed genes as defined in A. Gene lengths were rescaled to the same size as shown for the X-axis. (G) Average enrichment plots for NIPBL ChIP-seq signals in activated neutrophils, pretreated with or without transcription inhibitors for 2 h, plotted as a function of genomic distance from the center of TSSs (±2 kb), gated on differentially expressed genes as defined in A. (H) Average enrichment plots for NIPBL ChIP-seq signals in activated neutrophils, pretreated with or without transcription inhibitors for 2 h, plotted as a function of genomic distance from the center of TSSs (±2 kb). Genes were grouped according to expression levels in activated neutrophils as defined by GRO-seq signals.
To determine how upon calcium entry NIPBL accumulation was affected by transcription amplitude, genes were ranked based on GRO-seq reads and compared with the strength of NIPBL occupancy at TSSs. We found that the strength of DRB-accumulated NIPBL occupancy at promoters positively correlated with nascent transcript abundance (Fig. 4H). Specifically, interference with transcriptional elongation slightly lowered NIPBL occupancy at modestly transcribed genes whereas genes that were transcribed at relatively high rates showed a substantial increase in NIPBL binding strength (Fig. 4H). These data indicate that transcriptional elongation facilitates NIPBL occupancy and promotes NIPBL translocation as previously reported for cohesin (Busslinger et al. 2017). Taken together, these data indicate that upon calcium entry NIPBL is sequestered at active promoters and translocated coordinately with elongating RNA polymerase.
Targeting histone acetyltransferases, chromatin remodelers, and components of the transcriptional and cohesin machinery for acute depletion
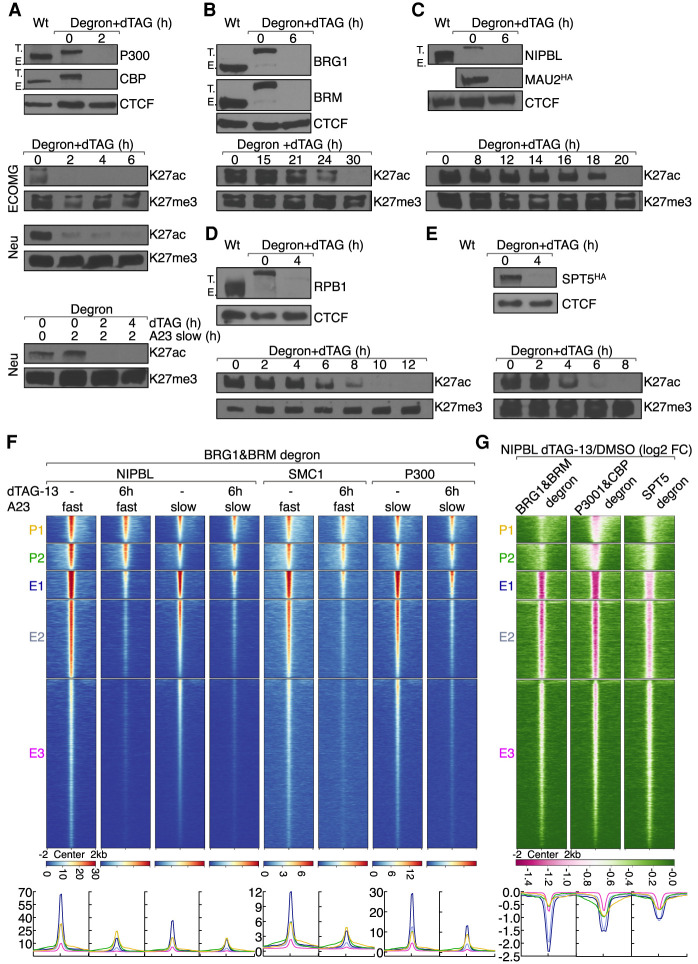
The data described above indicate that calcium signaling promotes P300, BRG1, and RNAPII occupancy at NIPBL-bound sites. To explore the immediate effects of these factors in orchestrating calcium-induced NIPBL occupancy we applied the FKBP12F36V acute degradation system. FKBP12F36V permits, in the presence of a heterobifunctional molecule named dTAG, rapid, specific, and near-complete degradation of targeted proteins (Nabet et al. 2018). The acute degradation system was established in ECOMG progenitor cells using CRISPR–Cas9 genome editing. Specifically, proteins of interest were tagged in frame at the N-terminal or C- terminal region with FKBP12F36V (Supplemental Fig. S5A). To monitor expression levels, tagged proteins were also marked with eYFP or mScarlet as well as an HA epitope (Supplemental Fig. S5A). As predicted, incubation with dTAG-13 rapidly lowered FKBP12F36V fusion protein fluorescence (Supplemental Fig. S5B). Western Blot validated these findings and showed near-complete loss of expression in response to incubation with dTAG-13 (Supplemental Fig. S5C,D). Incubation of wild-type ECOMG cells or neutrophils differentiated from ECOMG cells with dTAG-13 did not perturb cell viability, growth, or differentiation for up to 5–7 d of treatment (data not shown). Fusion with FKBP12F36V of targeted factors did not lead to constitutive, basal levels of degradation (data not shown). Basal protein abundance was equivalent in FKBP12F36V targeted cells when compared with parental lines cultured in the absence of dTAG-13 (data not shown). Additionally, targeted cells divided at similar rates and appeared indistinguishable from the parental cell lines cultured in the absence of dTAG-13. Thus, FKBP12F36V tagging does not abrogate essential neutrophil functions but permits, upon stimulation with dTAG-13, rapid and near complete depletion of targeted proteins of interest.
Histone acetyltransferases as well as transcription elongation are essential to maintain global H3K27ac abundance
The transcription coactivators P300 and CBP are highly homologous acetyltransferases that establish the H3K27ac and H3K18ac landscape (Jin et al. 2011; Raisner et al. 2018). As a first approach, to determine whether P300 and CBP directly regulate H3K27ac abundance, ECOMG cells harboring P300-FKBP12F36V or CBP-FKBP12F36V were cultured in the presence of dTAG-13. We found that neither P300 nor CBP were essential for viability, proliferation, and differentiation of ECOMG cells as well as maintenance of H3K27ac abundance (Supplemental Table S6; data not shown). These data raised the possibility that P300 and CBP act redundantly to maintain H3K27ac abundance. To examine this possibility, we generated ECOMG cells that harbored the FKBP12F36V degron in both the P300 and CBP loci. The resulting cell line is referred to as “P300&CBP degron.” FACS analysis showed that P300 protein levels remained equivalent whereas CBP protein abundance decreased during neutrophil differentiation (Supplemental Fig. S5B). Simultaneous degradation of P300 and CBP was tolerated for 6 h without obvious effects on cell viability (Supplemental Table S6). Culturing P300&CBP targeted cells for longer time periods (18 h), however, resulted in extensive cell death (Supplemental Table S6). We found that in resting and activated neutrophils within 2-h depletion of both P300 and CBP resulted in a global loss of H3K27ac abundance, while H3K27me3 levels remained unaffected (Fig. 5A). Thus, P300 and CBP maintain H3K27ac abundance across the enhancer landscape.
Figure 5.
Factors that directly orchestrate H3K27ac abundance and NIPBL occupancy. (A) The top panel shows Western blot validating dTAG-13 mediated depletion of P300 and CBP abundance in neutrophils expressing “P300&CBP degrons.” CTCF was probed to serve as a loading control. Note that successful biallelic tagging is shown by the higher-molecular-weight band (T.) and the lack of native-sized protein (E.) in CRISPR-modified cells. The bottom panels indicate Western blots showing changes in H3K27ac abundance upon dTAG-13-mediated P300 and CBP degradation. H3K27me3 was probed as a loading control. Whole-cell lysates were extracted from ECOMG cells, resting neutrophils, and activated neutrophils, respectively. (B) The top panel shows Western blot validating dTAG-13 mediated depletion of BRG1 and BRM abundance in neutrophils expressing “BRG1&BRM degrons.” Antibodies are indicated to the right. CTCF was probed to serve as a loading control. The bottom panel indicates Western blot showing showing changes in H3K27ac abundance upon dTAG-13-mediated P300 and CBP degradation. H3K27me3 was probed to serve as a loading control. Whole cell lysates were extracted from resting neutrophils. (C) As in B for NIPBL and MAU2 in “NIPBL&MAU2 degron” neutrophils. Note that MAU2 was blotted with anti-HA antibody. (D) As in B for RPB1 in “RPB1 degron” neutrophils. (E) As in B for SPT5 in “SPT5 degron” neutrophils. Note that SPT5 was blotted with anti-HA antibody. (F) Heat map of NIPBL, SMC1, and P300 ChIP-seq signals in activated neutrophils in the presence or absence of dTAG-13, plotted for five clusters of genomic regions defined in Figure 2A. Heat map was generated by gating on the maximum signal intensity of NIPBL occupancy using A23187 slow activation conditions. Below the heat map corresponding average enrichment plots are shown. Near-complete degradation was achieved by 6 h. Therefore, 6 h of dTAG-13 treatment was chosen to study the immediate consequences for loss of BRG1&BRM expression while minimizing indirect effects on transcription and genome organization caused by cell death. (G) Heat map of log2 FC of NIPBL ChIP-seq signals in dTAG-13 versus DMSO-treated activated neutrophils plotted for five clusters of regions defined in Figure 2A. Heat map was sorted by the minimum signal intensity of log2 FC of NIPBL in “P300&CBP degron” cells. Pink color indicates signal loss upon dTAG-13 treatment. Below the heat map is indicated the corresponding average enrichment plots with Y-axis values indicated as log2FC. Below the heat map are corresponding average enrichment plots. “BRG1&BRM degron,” “P300&CBP degron,” and “SPT5 degron” neutrophils were pretreated with dTAG-13 for 6, 2, and 4 h before activation, respectively.
The chromatin remodeler BAF complexes are composed of a core ATPase, BRG1 or BRM, and ∼15 associated subunits (Alfert et al. 2019). BRG1 and BRM are >75% identical and display similar activities in vitro (Phelan et al. 1999; Gatchalian et al. 2020). To explore whether BRG1 and BRM regulate H3K27ac abundance and NIPBL-binding directly, we began by degrading them individually. We found that BRG1 and BRM were dispensable for cell viability, proliferation, differentiation, and H3K27ac maintenance (Supplemental Table S6; data not shown). To determine whether they act redundantly we generated ECOMG cells that harbored the FKBP12F36Vdegron in both the BRG1 and BRM loci. FACS analysis showed that during neutrophil differentiation BRM protein levels remained unaffected whereas BRG1 protein levels decreased (Supplemental Fig. S5B). Depletion of both BRG1 and BRM protein levels was tolerated for 24 h without obvious cell death, but we found that when incubated for 36 h the majority of cells had died (Supplemental Table S6). Near complete degradation of BRG1 and BRM was achieved by 6 h (Fig. 5B). Only prolonged depletion of both BRG1 and BRM modestly decreased H3K27ac levels, immediately prior to massive cell death, indicating that BRG1 and BRM do not directly modulate H3K27ac abundance (Fig. 5B).
To explore whether nascent transcription, plausibly involving lncRNAs or eRNAs, regulates H3K27ac abundance, we tagged the largest subunit of RNAPII, RPB1, as well as SPT5, an essential part of the RNAPII elongation complex with the FKBP12F36V-degron (Zhou et al. 2012; Shetty et al. 2017). We found that during neutrophil differentiation RPB1 and SPT5 protein levels substantially decreased (Supplemental Fig. S5B). Degradation of RPB1 or SPT5 was tolerated for 8 h without obvious cell death whereas by 18 h the majority of cells had died (Supplemental Table S6). Near-complete degradation of RBP1 and SPT5 was achieved by 4 h. We found that depletion of either RPB1 or SPT5 resulted in global loss of H3K27ac abundance indicating that transcriptional elongation is essential to maintain H3K27ac abundance (Fig. 5D,E).
To determine whether NIPBL and MAU2 expression is essential to maintain H3K27ac abundance, differentiated ECOMG cells expressing NIPBL-FKBP12F36V-EYFP or MAU2-FKBP12F36V-mScarlet were cultured in the presence of dTAG-13. We found that neither NIBPL nor MAU2 expression alone was required for ECOMG cell and neutrophil viability, proliferation, differentiation, and H3K27ac maintenance (Supplemental Table S6). To determine whether they act redundantly, we generated ECOMG cells that express both NIPBL-FKBP12F36V and MAU2-FKBP12F36V. Degradation of both NIPBL and MAU2 was tolerated for 18 h without obvious cell death, but by 24 h the majority of cells had died (Supplemental Table S6). Near-complete depletion of NIPBL and MAU2 abundance was achieved by 6 h of culturing ECOMG cells in dTAG-13. Only prolonged NIPBL and MAU2 depletion (18–20 h) showed decreased H3K27ac levels indicating that as expected NIPBL and MAU2 do not modulate H3K27ac abundance directly (Fig. 5C). Taken together, these data indicate that P300 and CBP as well as transcriptional elongation maintain H3K27ac abundance.
NIPBL occupancy at enhancers vs. promoters is differentially regulated by chromatin remodelers, histone acetyltransferases, and transcriptional elongation
The data described above indicate that calcium-induced recruitment of BRG1 and NIPBL at promoters occurs with relatively slow kinetics (3–4 h) when compared with enhancers. To determine whether the differences in NIPBL occupancy at enhancers reflect differences in occupancy of chromatin remodelers “BRG1&BRM degron” neutrophils were activated and cultured in the absence or presence of dTAG-13 (Fig. 5F). We found that NIPBL occupancy at enhancers was severely perturbed upon depleting BRG1 and BRM expression (Fig. 5F; Supplemental Fig. S6A). In contrast, NIPBL occupancy at promoter regions was only marginally affected upon eliminating BRG1 and BRM expression (Fig. 5F). Likewise, while depleting BRG1 and BRM levels greatly reduced SMC1 occupancy at active enhancers SMC1 occupancy at promoters was only marginally affected (Fig. 5F; Supplemental Fig. S6A). To determine whether P300 occupancy is regulated by BRG1 and BRM we examined “BRG1&BRM degron” neutrophils for P300 occupancy. We found that activated neutrophils depleted for BRG1 and BRM expression showed significant lower levels of P300 occupancy at enhancers but not at promoters (Fig. 5F; Supplemental Fig. S6A).
To determine whether P300 and CBP expression facilitates calcium-induced NIPBL occupancy, activated “P300&CBP degron” neutrophils were cultured in the absence or presence of dTAG-13 and examined for NIPBL occupancy (Fig. 5G). We found that calcium-induced NIPBL occupancy was substantially lowered at active enhancers in cells depleted for P300 and CBP (Fig. 5G; Supplemental Fig. S6A). Conversely, NIPBL occupancy at promoters was only marginally affected by depleting P300 and CBP expression (Fig. 5G; Supplemental Fig. S6A). To determine whether transcriptional elongation instructs calcium-induced NIPBL occupancy, activated neutrophils depleted for SPT5 expression were examined for NIPBL occupancy. We found that calcium-induced NIPBL occupancy was substantially lowered at active enhancers in cells depleted for SPT5 while NIPBL occupancy at promoters was only slightly affected (Fig. 5G; Supplemental Fig. S6A). Taken together, these observations indicate that chromatin remodelers, histone acetyltransferases and transcriptional elongation, facilitate NIPBL and SMC1 occupancy at enhancers rather than at promoters.
Neutrophil differentiation is closely associated with a decline in loop extrusion
Our previous observations as well as the data described here indicate that neutrophil differentiation is accompanied by large-scale changes in nuclear architecture (Zhu et al. 2017; Denholtz et al. 2020). These findings raised the question as to whether these alterations in genome topology are established by changes in the expression of genes encoding for NIPBL, MAU2, CTCF, and RAD21. To address this question, we probed the expression of NIPBL, MAU2, CTCF, and RAD21 using ECOMG cell lines that carry fluorescently labeled degrons. Briefly, ECOMG cells expressing NIPBL-FKBP12F36V-EYFP, MAU2-FKBP12F36V-mScarlet, CTCF-FKBP12F36V-EYFP, and RAD21-FKBP12-F36V-EYFP were differentiated and analyzed for EYFP or mScarlet expression. Interestingly, we found that NIPBL, CTCF, MAU2, and RAD21 expression all substantially declined in differentiating neutrophils (Supplemental Fig. S5B). Taken together, these data suggest that the large-scale alterations in nuclear architecture associated with neutrophil differentiation is dictated by alterations in the expression of genes encoding for factors that instruct loop extrusion.
Calcium-induced transcription is instructed by RAD21, chromatin remodelers, and histone acetyltransferases
To determine whether CTCF, RAD21, and WAPL, instruct calcium-induced NIPBL occupancy, activated neutrophils carrying degrons for these factors were cultured in the absence or presence of dTAG-13 and examined for NIPBL occupancy. As predicted, calcium-induced NIPBL occupancy was not lowered at active enhancers and promoters in cells depleted for CTCF, RAD21, and WAPL (unpublished observations). To determine whether and how depletion of RAD21, BRG1, BRM, P300, and CBP modulate calcium-induced transcription, activated neutrophils depleted for these factors were analyzed for RPB1 occupancy. We found that upon calcium entry depletion of RAD21, P300, and CBP as well as BRG1 and BRM lowered RBP1 occupancy (Supplemental Fig. S6B). Taken together, these data indicate that in activated neutrophils calcium-induced transcription is instructed by histone acetyltransferases, chromatin remodelers and the cohesin machinery.
Calcium signaling activates NFAT to promote NIPBL occupancy at enhancers
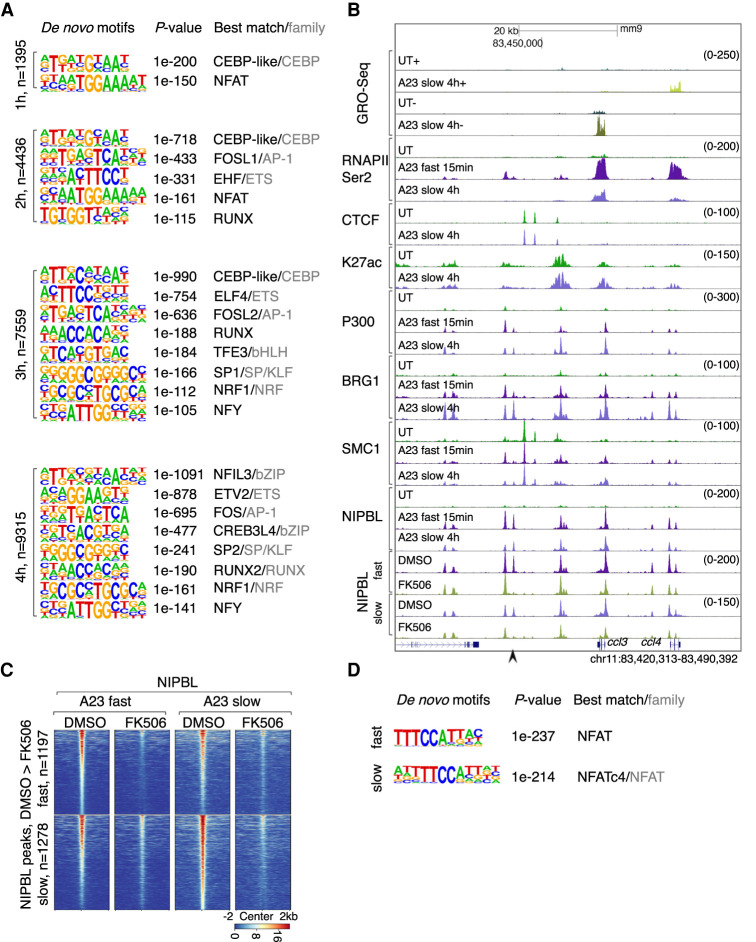
To identify potential TFs that promote calcium-signaling induced NIPBL occupancy, we examined induced NIPBL-binding sites for enriched DNA sequences. A progressive enrichment of TF motifs across the activation time course was identified. At 1 h after activation, induced NIPBL-binding sites were enriched for cis elements associated with CEBP and NFAT occupancy (Fig. 6A; Supplemental Fig. S7). Calcium signaling for a duration of 2 h doubled the number of NIPBL-binding sites that were enriched for DNA sequences associated with NFAT, CEBP, ETS, AP-1, and RUNX occupancy (Fig. 6A; Supplemental Fig. S7). The number of NIPBL-bound sites increased 3 h after activation and were enriched for cis elements associated with bHLH, SP/KLF, NRF, and NFY occupancy (Fig. 6A; Supplemental Fig. S7). The number of NIPBL-bound sites continued to increase 4 h after activation, but the composition of TF binding motifs essentially remained the same (Fig. 6A; Supplemental Fig. S7). Notably, however, cis elements associated with calcium-induced NIPBL-occupancy at promoters were distinct from those associated with enhancers. Specifically, we found that calcium-induced NIPBL occupancy at promoters was enriched for GC-rich DNA sequences (SP/KLF) and appeared with different kinetics following calcium entry when compared with enhancers (3-4 h) (Supplemental Fig. S7).
Figure 6.
Calcium signaling activates NFAT to promote NIPBL occupancy. (A) Motifs enriched in the vicinity of NIPBL-binding sites (±200 bp) that are up-regulated in activated neutrophils at respective time points using GC-matched genomic backgrounds. Note that all members of a TF family tended to show similar enrichment due to motif similarity. P-value for motif enrichment, the best-matched TF and TF family if applicable, are indicated at the right of the sequence logos (cutoff: P < 1 × 10−100). (B) Genome browser tracks depicting GRO-seq and ChIP-seq reads in untreated and activated neutrophils, cultured in the presence or absence of FK506. A representative locus mmp8 is shown. Black arrowheads indicate NIPBL-bound sites that were down-regulated in FK506-treated neutrophils. (C) Heat maps of NIPBL occupancy in activated neutrophils cultured in the absence or presence of FK506, using a window of ± 2 kb centered on NIPBL bound sites. Heat maps were gated on down-regulated NIPBL-bound sites identified in FK506-treated versus DMSO-treated activated neutrophils. Corresponding average enrichment plots are shown below. (D) Motifs enriched in the vicinity of NIPBL-bound sites (±200 bp) that were depleted in FK506-treated versus DMSO-treated activated neutrophils. A GC-matched genomic background was used for the analysis. P-value for motif enrichment, the best-matched TF and TF family if applicable are indicated at the right of the sequence logos (cutoff: P < 1 × 10−100).
To validate these findings, we tested whether NFAT binding sites were associated with calcium-induced NIPBL occupancy. Specifically, activated neutrophils were cultured in the absence or presence of FK506, a well-characterized NFAT inhibitor and examined for NIPBL binding. As predicted, inhibition of NFAT activity interfered with calcium-induced NIPBL occupancy (Fig. 6B,C). Specifically, pretreatment with FK506 interfered with calcium-induced NIPBL occupancy at 1197 sites (fast activation) and at 1278 sites (slow activation) (Fig. 6C). Consistent with these results, de novo motif analysis revealed that calcium-responsive FK506-sensitive NIPBL-binding sites were predominantly enriched for NFAT motifs (Fig. 6D). Taken together, these data indicate that upon calcium influx NIPBL is initially recruited at enhancers that are enriched for NFAT, PU.1, CEBP, and AP-1 binding sites while at later stages NIPBL is sequestered at promoters characterized by GC-rich DNA sequences.
PU.1 modulates calcium-induced NIPBL occupancy across the chromatin landscape
It is well established that PU.1 is a pioneering factor that orchestrates neutrophil development and activation (Anderson et al. 1998; Iwasaki et al. 2005; Fischer et al. 2019; Minderjahn et al. 2020). The data described above raised the possibility that PU.1 as a pioneering factor acts to promote NIPBL occupancy. To determine whether, indeed, PU.1 expression is essential to instruct NIPBL occupancy we generated PU.1-knockout (KO) ECOMG-derived neutrophils. ECOMG-derived PU.1 wild-type (WT) and knock-out (KO) neutrophils were stimulated with A23187 and examined for calcium-induced NIPBL occupancy. We found that depletion of PU.1 globally modulated NIPBL binding (Supplemental Fig. S8A,B). Specifically, PU.1 depletion resulted in a loss of 4780 NIPBL-bound sites and a gain of 11434 NIPBL-bound sites (Supplemental Fig. S8B). Likewise, we found that PU.1 depletion coordinately changed SMC1 occupancy (Supplemental Fig. S8B). To determine whether upon depletion of PU.1 expression loss or gain of NIPBL occupancy were differentially associated with cis elements we performed a de novo motif analysis. We found that upon depleting PU.1 abundance, calcium-induced loss of NIPBL binding sites were enriched for DNA sequences associated with PU.1 binding, while calcium-induced gain of NIPBL binding sites were enriched for DNA sequences associated with CEBP binding, plausibly involving a competitive displacement mechanism (Cloutier et al. 2009; Hosokawa et al. 2018). Taken together, these data imply that PU.1 uses distinct mechanisms to direct NIPBL occupancy across the chromatin landscape.
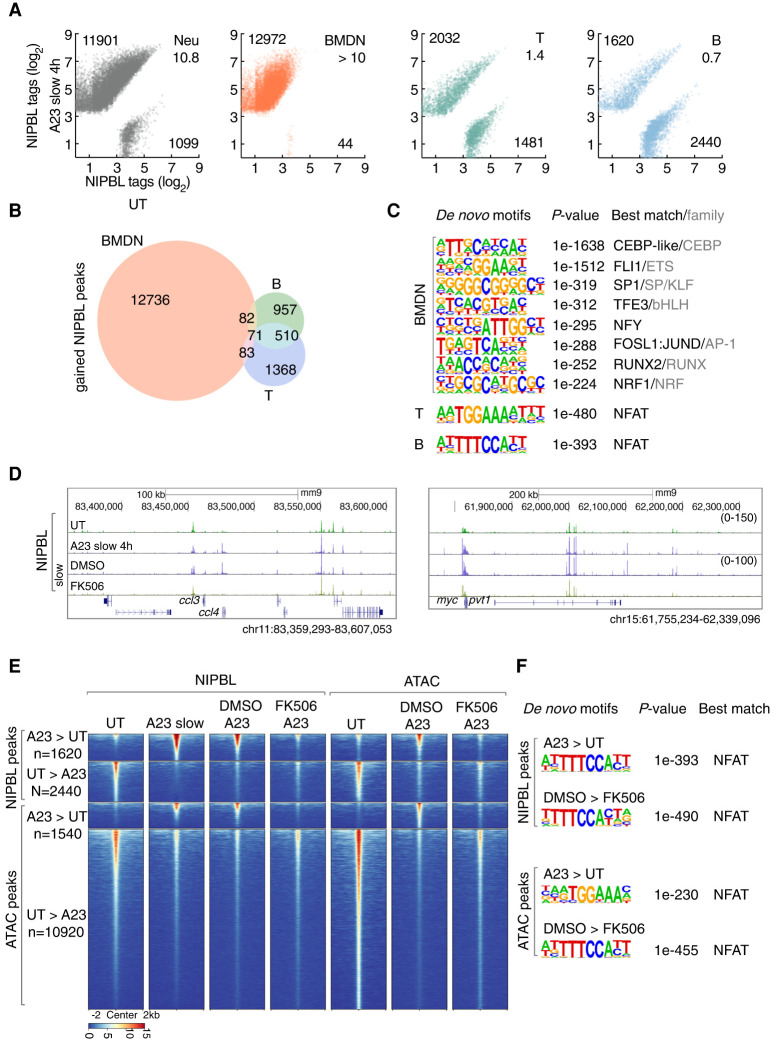
Calcium signaling instructs lineage-specific NIPBL occupancy in primary immune cells
The data described above indicate that calcium signaling induces NIPBL recruitment to active enhancers and promoters in neutrophils. However, these experiments were performed using in vitro differentiated neutrophils that were derived from ECOMG cells. To validate these findings for primary cells, neutrophils were isolated from the bone marrow and activated with A23187. We found that calcium influx in primary bone marrow-derived neutrophils (BMDN) rapidly modulated NIPBL-binding with the majority of binding sites showing increased NIPBL occupancy (Fig. 7A). We next examined NIPBL-binding in A23187-activated primary T and B cells isolated from the spleen. We found that calcium influx in primary T cells revealed equal ratios of elevated and declining levels of NIPBL occupancy (Fig. 7A). Calcium signaling in primary B cells showed a reverse pattern with a substantial fraction of NIPBL-bound sites being associated with decreased occupancy (Fig. 7A). Calcium-induced NIPBL bound sites showed substantial overlap between activated B and T lineage cells (Fig. 7B). In contrast, we found that only a minor fraction of calcium-induced NIPBL-binding sites overlapped between BMDN versus T and B cells (Fig. 7B). Calcium-induced NIPBL-binding sites in BMDN cells were enriched for a wide spectrum of cis elements, resembling that of ECOMG-derived neutrophils, whereas T and B cells were predominantly enriched for NFAT binding sites (Fig. 7C). In line with these results, we found FK506 severely impaired enrichment of NIPBL occupancy for a cluster of genes encoding for chemokines as well as a genomic region spanning the c-myc and pvt1 loci (Fig. 7D). To determine whether activation of NFAT promotes chromatin remodeling at calcium-induced NIPBL-bound sites we performed ATAC-seq. We found that activated-B cells were associated with altered chromatin accessibility at calcium-induced NIPBL bound sites as well as calcium-repressed NIPBL-bound sites (Fig. 7E). Notably, we found that NFAT inhibition by FK506 pretreatment abolished the increase in chromatin accessibility but only modestly reversed the decrease in chromatin accessibility (Fig. 7E). Finally, de novo motif analysis revealed that FK506-down-regulated ATAC-seq reads were enriched for NFAT cis elements suggesting that NFAT binds these sites directly to promote chromatin accessibility and to facilitate NIPBL occupancy (Fig. 7F). Collectively, these data indicate that calcium signaling orchestrates a lineage-specific pattern of NIPBL occupancy in primary immune cells and that NFAT plays a key role in modulating chromatin accessibility at NIPBL bound sites.
Figure 7.
Calcium influx in primary immune cells modulates NIPBL occupancy. (A) Scatter plot for log2 tag counts for NIPBL peaks in untreated versus activated ECOMG-derived neutrophils, bone marrow-derived neutrophils (BMDN), primary T and B cells, respectively, assessed for a 200-bp region centered on differential NIPBL-bound sites. Numbers of gained and lost NIPBL-bound sites (FC > 4) and ratios between them are indicated. (B) Venn diagram showing overlap and specificity of gained NIPBL-binding sites in activated BMDN, primary T cells and B cells. Peaks were considered overlapping if the distance separating peak centers were <100 bp. Numbers of specific and overlapping bound sites are indicated for each group. (C) Motifs enriched in the vicinity of NIPBL-binding sites (± 200 bp) that were gained in activated BMDN, primary T cells and B cells using GC-matched genomic backgrounds, respectively. P-values for motif enrichment, the best-matched TF and TF family if applicable are indicated at the right of the sequence logos (cutoff: P < 1×10−100). (D) Genome browser tracks of NIPBL-bound sites derived from untreated and activated primary B cells cultured in the presence or absence of FK506 at representative loci, ccl4 and myc. (E) Heat map of NIPBL-bound sites and ATAC-seq signals in untreated and activated primary B cells cultured in the presence or absence of FK506, across genomic regions that were centered (±2 kb) on NIPBL-bound sites. (F) The top panels indicate motifs enriched within the vicinity of NIPBL-bound sites (±200 bp) that are gained in activated primary B cells using GC-matched genomic backgrounds. The bottom panels show motifs enriched in the vicinity of accessible chromatin regions defined by ATAC-seq (±200 bp) that were lost in activated primary B cells cultured in the presence of FK506 using GC-matched genomic backgrounds. P-values for motif enrichment, the best-matched TF and TF family if applicable, are indicated to the right of the sequence logos (cutoff: P < 1 × 10−100).
Discussion
Previous studies demonstrated that the transition from progenitors to differentiated neutrophils, in mice and humans, is closely associated with large-scale changes in genome topology (Zhu et al. 2017; Denholtz et al. 2020). These changes involve a distinct pattern of remote genomic interactions that span entire chromosomes and a uniquely fragmented compartment structure (Zhu et al. 2017; Denholtz et al. 2020). During neutrophil differentiation how are these alterations in long-range genomic interactions and compartmentalization established? We suggest that they are instructed by a decrease in loop extrusion. This notion is supported by our observation that RAD21, NIPBL, and MAU2 expression levels readily decline during the transition from progenitors to terminally differentiated neutrophils. These findings raise the question of whether and how the decrease in NIPBL, MAU2, and RAD21 expression in differentiating neutrophils is linked to the alterations in nuclear architecture. The decline in NIPBL, MAU2, and RAD21 expression during neutrophil differentiation instructs hyper-fragmented compartmentalization that mimics the increase in compartment segregation observed in NIPBL-deficient hepatocytes (Schwarzer et al. 2017). Thus, we suggest that the decline in NIPBL, MAU2, and RAD21 expression during neutrophil differentiation instructs a distinct chromatin folding pattern that characterizes, at least in part, the terminally differentiated hyper-fragmented neutrophil compartment structure.
While resting neutrophils are associated with fragmented compartmentalization, upon calcium-mediated activation the neutrophil genome adopts a nuclear architecture that resembles that of other immune cells, essentially reversing hyper-fragmented compartmentalization. We found that these changes in nuclear architecture are associated with a global and rapid increase in NIPBL and SMC1 occupancy. Thus, upon calcium entry elevated levels of NIPBL dictate a nuclear architecture that is akin to that of other immune cells in terms of compartmentalization. This then raises the question why hyper-fragmented compartments are established in terminally differentiated neutrophils? We suggest that prior to activation a highly fragmented compartment structure enforces efficient insulation of distant regulatory elements to prevent stochastic activation of a neutrophil specific gene program. Conversely, upon calcium entry loss of highly segregated compartmentalization may facilitate local genomic interactions to bring into close spatial proximity enhancer and promoter elements allowing the expression of genes associated with an activated neutrophil specific gene program. Consistent with such a scenario are our observations that depletion of RAD21 in differentiated neutrophils altered calcium-induced RNA polymerase II occupancy. We note that gene expression modulated by the cohesin machinery is not unique to activated neutrophils. Recent studies indicated that cohesin expression is required to induce the expression of an inflammatory gene program in macrophages (Cuartero et al. 2018). Thus, loop extrusion-induced alterations in nuclear architecture might be a shared mechanism that is used by macrophages and neutrophils to orchestrate inducible programs of gene expression.
How does calcium signaling recruits NIPBL occupancy with such great speed and precision across the chromatin landscape? Calcium-induced NIPBL occupancy at enhancers involves a wide spectrum of TFs. Prominent amongst these are TFs that regulate gene expression in response to a spectrum of stimuli, including NFAT and AP1 as well as TFs that orchestrate lineage-specific gene programs such as PU.1 and CEBP. The second step involves TF-dependent recruitment of the histone acetyltransferases P300 and CBP. Once recruited P300 and CBP establish a calcium-induced specific H3K27ac landscape. Likewise, the chromatin remodelers BRG1 and BRM are sequestered at NIPBL-bound sites. Whether the recruitment of P300 and CBP as well as BRG1 and BRM engage sequential or parallel pathways or pathways that involve complex feedback mechanisms, remains to be determined, but it is evident that they are essential to sequester NIPBL to its target sites. In addition to histone acetyltransferases and chromatin remodelers we found that transcriptional elongation plays a role as well in elevating H3K27 abundance and targeting NIPBL to enhancers. How may transcriptional elongation modulate H3K27ac levels and facilitate NIPBL occupancy? We suggest that noncoding transcripts, lncRNAs and/or eRNAs, function to modulate the catalytic activity of P300 and CBP. Finally, it is conceivable that noncoding transcription facilitates the assembly of hubs or condensates to facilitate NIPBL occupancy.
While depletion of BRG1 and BRM severely impaired calcium-induced NIPBL occupancy at enhancers, NIPBL binding at promoters was only modestly affected. How does recruitment of NIPBL to promoter regions differ from that to enhancers? We found that distinct cis elements are associated with NIPBL occupancy at promoters versus enhancers. Induced NIPBL-binding sites at promoters are enriched for DNA sequences associated with SP/KLF binding sites (GC-rich) rather than NFAT, AP-1, PU.1, and CEBP motifs that are associated with enhancers. The enrichment for SP/KLF cis elements is particularly intriguing since SP1 consensus DNA sequences have been shown to be associated with noncanonical secondary DNA structures such as G-quadruplexes (G4) (Raiber et al. 2012). We validated these observations using a bioinformatics approach, namely Pqsfinder. Notably, we found that indeed G4 DNA structures were enriched at induced NIPBL-binding sites (data not shown) (Hon et al. 2017). G4 structural motifs have also been linked to transcription and are enriched at promoters and 5′ UTRs associated with highly transcribed genes (Hänsel-Hertsch et al. 2016, 2018). While chromatin remodelers do not appear to promote NIPBL occupancy at promoters, the histone acetyltransferases P300 and CBP also modulate NIPBL occupancy at promoters, albeit to a much lesser degree. Thus, noncanonical DNA structures across the promoter landscape may permit recruitment of NIPBL in a pathway that is independent of chromatin remodelers but may involve the activities of histone acetyltransferases. Finally, we note that it is conceivable that in order to enable NIPBL occupancy at promoters, GC-rich noncanonical secondary DNA structures may enrich for chromatin accessible regions that resemble chromatin remodeling-induced nucleosome depleted regions at enhancers. In sum, while chromatin remodelers are key to facilitate NIPBL occupancy at enhancers, promoters use noncanonical GC-rich DNA structures to sequester NIPBL.
While much remains to be learned, a cardinal conclusion reached from the studies described here is that transcription factors, noncoding transcription, the deposition of H3K27ac, and depletion of nucleosomes are instrumental in sequestering the cohesin machinery across the active enhancer repertoire.
Materials and methods
Cell culture
In vitro differentiated neutrophils from ECOMG cells were used as a model system to study nuclear architecture in response to calcium influx in neutrophils. ECOMG cells were cultured and differentiated as previously described (Zhu et al. 2017). For inhibitors, neutrophils were treated with 10 µM Triptolide (Cayman), 100 µM 5,6-dichloro-1-β-D-ribofuranosyl-1H-benzimidazole (Cayman) or 10 µM flavopiridol (Cayman) for 30 min, or 2 µM FK506 for 2 h before activation. Primary B cells were treated with 2 µM FK506 (Cayman) for 2 h before activation. Inhibitors were kept in culture during activation time course. Acute degradation was induced by treating the cells with 0.5 µM dTAG-13 (Tocris) before activation. dTAG-13 was kept in culture during activation time course. The time series degradation experiments were performed by inducing protein degradation at the beginning of the time course and harvesting the samples at different time points. RPMI-1640 (Gibco) contains 0.42 mM calcium. Additional CaCl2 was supplied to the medium before activation to a final concentration of 1 mM. A23187 was purchased from Sigma and Cayman. Neutrophils were activated either using fast activation conditions (20 µM A23187 for 15 min), or using slow activation conditions (5 µM A23187 for 4 h). These two activation conditions were used throughout the manuscript unless otherwise mentioned.
Data availability
Data sets generated in this study are available as a series in the GEO database under accession number GSE154051.
Acknowledgments
Sequencing was performed at the IGM Genomics Center, University of California at San Diego, La Jolla, CA, supported by grant number P30CA023100. Imaging was performed at the microscopy core of the School of Medicine, University of California at San Diego, La Jolla, CA (CA23187100 and NS047101). This study was supported by grants from the National Institutes of Health to C.M. (AI00880, AI09599, and AI102853).
Author contributions: Y.Z., M.D., and C.M. conceived this project. Y.Z. designed and performed all experiments. H.L. cloned repair template targeting cbp gene locus, isolated primary T and B cells from mice and helped with FACS analysis. Y.Z. analyzed all data. H.L. analyzed compartment strength. Y.Z. and C.M. prepared the manuscript with input from other authors. C.M. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.343475.120.
References
- Alfert A, Moreno N, Kerl K. 2019. The BAF complex in development and disease. Epigenetics and Chromatin 12: 19 10.1186/s13072-019-0264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Smith KA, Pio F, Torbett BE, Maki RA. 1998. Neutrophils deficient in PU.1 do not terminally differentiate or become functionally competent. Blood 92: 1576–1585. 10.1182/blood.V92.5.1576 [DOI] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303: 1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, Van Der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507. 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254. 10.1016/S1097-2765(00)80420-7 [DOI] [PubMed] [Google Scholar]
- Cloutier A, Guindi C, Larivée P, Dubois CM, Amrani A, McDonald PP. 2009. Inflammatory cytokine production by human neutrophils involves C/EBP transcription factors. J Immunol 182: 563–571. 10.4049/jimmunol.182.1.563 [DOI] [PubMed] [Google Scholar]
- Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Robles-Rebollo I, Xiao X, Wang YF, Barozzi I, et al. 2018. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol 19: 932–941. 10.1038/s41590-018-0184-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Zhu Y, He Z, Lu H, Isoda T, Döhrmann S, Nizet V, Murre C. 2020. Upon microbial challenge, human neutrophils undergo rapid changes in nuclear architecture and chromatin folding to orchestrate an immediate inflammatory gene program. Genes Dev 34: 149–165. 10.1101/gad.333708.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Walter C, Tönges A, Aleth H, Jordão MJC, Leddin M, Gröning V, Erdmann T, Lenz G, Roth J, et al. 2019. Safeguard function of PU.1 shapes the inflammatory epigenome of neutrophils. Nat Immunol 20: 546–558. 10.1038/s41590-019-0343-z [DOI] [PubMed] [Google Scholar]
- Fric J, Zelante T, Wong AYW, Mertes A, Yu HB, Ricciardi-Castagnoli P. 2012. NFAT control of innate immunity. Blood 120: 1380–1389. 10.1182/blood-2012-02-404475 [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Liao J, Maxwell MB, Hargreaves DC. 2020. Control of stimulus-dependent responses in macrophages by SWI/SNF chromatin remodeling complexes. Trends Immunol 41: 126–140. 10.1016/j.it.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782. 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JH, Rowland BD. 2017. Cohesin: building loops, but not compartments. EMBO J 36: 3549–3551. 10.15252/embj.201798654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. 2017. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707.e14. 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, et al. 2016. G-quadruplex structures mark human regulatory chromatin. Nat Genet 48: 1267–1272. 10.1038/ng.3662 [DOI] [PubMed] [Google Scholar]
- Hänsel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. 2018. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc 13: 551–564. 10.1038/nprot.2017.150 [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 17: 2205–2232. 10.1101/gad.1102703 [DOI] [PubMed] [Google Scholar]
- Hogan PG, Lewis RS, Rao A. 2010. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol 28: 491–533. 10.1146/annurev.immunol.021908.132550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon J, Martínek T, Zendulka J, Lexa M. 2017. Pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics 33: 3373–3379. 10.1093/bioinformatics/btx413 [DOI] [PubMed] [Google Scholar]
- Hosokawa H, Ungerbäck J, Wang X, Matsumoto M, Nakayama KI, Cohen SM, Tanaka T, Rothenberg EV. 2018. Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity 48: 1119–1134.e7. 10.1016/j.immuni.2018.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106: 1590–1600. 10.1182/blood-2005-03-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SYR, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30: 249–262. 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, Van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EF, Herzig A, Krüger R, Muth A, Mondal S, Thompson PR, Brinkmann V, von Bernuth H, Zychlinsky A. 2017. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife 6: 1–21. 10.7554/eLife.24437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967. 10.1016/j.cell.2006.09.040 [DOI] [PubMed] [Google Scholar]
- Ley K, Hoffman HM, Kubes P, Cassatella MA, Zychlinsky A, Hedrick CC, Catz SD. 2018. Neutrophils: new insights and open questions. Sci Immunol 3: eaat4579 10.1126/sciimmunol.aat4579 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minderjahn J, Schmidt A, Fuchs A, Schill R, Raithel J, Babina M, Schmidl C, Gebhard C, Schmidhofer S, Mendes K, et al. 2020. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU.1. Nat Commun 11: 402 10.1038/s41467-019-13960-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Uhlmann F. 2014. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505: 367–371. 10.1038/nature12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al. 2018. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14: 431–441. 10.1038/s41589-018-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. 2009. Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115: E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti I, Diab F, Gil SR, Mulugeta E, Casa V, Berutti R, Brouwer RWW, Dupé V, Eckhold J, Graf E, et al. 2020. MAU2 and NIPBL variants impair the heterodimerization of the cohesin loader subunits and cause Cornelia de Lange syndrome. Cell Rep 31: 107647 10.1016/j.celrep.2020.107647 [DOI] [PubMed] [Google Scholar]
- Petela NJ, Gligoris TG, Metson J, Lee BG, Voulgaris M, Hu B, Kikuchi S, Chapard C, Chen W, Rajendra E, et al. 2018. Scc2 Is a potent activator of cohesin's ATPase that promotes loading by binding Scc1 without Pds5. Mol Cell 70: 1134–1148.e7. 10.1016/j.molcel.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 3: 247–253. 10.1016/S1097-2765(00)80315-9 [DOI] [PubMed] [Google Scholar]
- Raiber EA, Kranaster R, Lam E, Nikan M, Balasubramanian S. 2012. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res 40: 1499–1508. 10.1093/nar/gkr882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, Haverty PM, Bainer R, Cheung T, Arnott D, et al. 2018. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep 24: 1722–1729. 10.1016/j.celrep.2018.07.041 [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. 2018. Organizational principles of 3D genome architecture. Nat Rev Genet 19: 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Kallgren SP, Demel C, Maier KC, Spatt D, Alver BH, Cramer P, Park PJ, Winston F. 2017. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol Cell 66: 77–88.e5. 10.1016/j.molcel.2017.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecchio C, Micheletti A, Cassatella MA. 2014. Neutrophil-derived cytokines: facts beyond expression. Front Immunol 5: 1–7. 10.3389/fimmu.2014.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaeth M, Feske S, Macian F. 2018. NFAT control of immune function: new frontiers for an abiding trooper. F1000Res 7: 260 10.12688/f1000research.13426.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. 2018. The energetics and physiological impact of cohesin extrusion. Cell 173: 1165–1178.e20. 10.1016/j.cell.2018.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143. 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Gong K, Denholtz M, Chandra V, Kamps MP, Alber F, Murre C. 2017. Comprehensive characterization of neutrophil genome topology. Genes Dev 31: 141–153. 10.1101/gad.293910.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sets generated in this study are available as a series in the GEO database under accession number GSE154051.