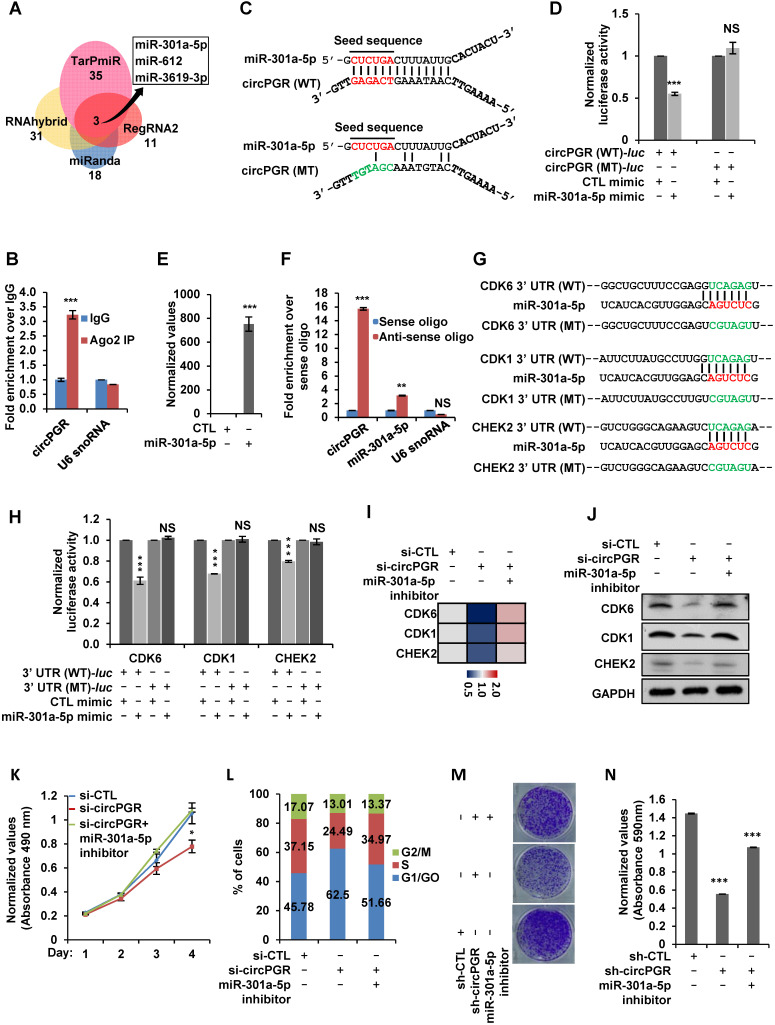
Figure 6.
CircPGR sponges miR-301a-5p to regulate the expression of multiple cell cycle genes and cell cycle progression. (A) Four different miRNA prediction algorithms as indicted were used to predict potential miRNAs that can bind to circPGR, and the highly confident miRNAs predicted were then overlapped, resulting in three miRNAs, miR-301a-5p, miR-612 and miR-3619-3p. (B) RNA immunoprecipitation (RNA IP) was performed in MCF7 cells stably expressing 3 × Flag-tagged AGO2 by using control IgG or anti-Flag antibody followed by RT-qPCR to examine the binding of circPGR and U6 snoRNA (±s.e.m., ***P < 0.001). (C) Sequence match between miR-301a-5p and linear sequence of circPGR (circPGR (WT)) or circPGR mutant form with the potential miR-301a-5p binding site mutated (circPGR (MT)) was shown. Seed sequence was highlighted in red in miR-301a-5p. (D) Full length linear sequence of circPGR (circPGR (WT)) as well as its mutant form with the potential miR-301a-5p binding site mutated (circPGR (MT)) were cloned into a luciferase reporter vector, which were then transfected into HEK293T cells with control mimic or miR-301a-5p mimic followed by luciferase activity measurement (±s.e.m., ***P < 0.001, NS: non-significant). (E) The expression of miR-301a-5p mimic was detected by stem-loop RT-qPCR (±s.e.m., ***P < 0.001). (F) Oligonucleotide pull-down assay was performed by incubated MCF7 cell lysates with sense or anti-sense oligonucleotide targeting circPGR followed by RT-qPCR to detect the associated RNA as indicated (±s.e.m., **P < 0.01, ***P < 0.001, NS: non-significant). (G) Sequence match between miR-301a-5p and 3' untranslated region (UTR) of CDK6, CDK1 or CHEK2 (3' UTR (WT)) as well as the corresponding mutant form with the predicted miR-301a-5p binding site mutated (3' UTR (MT)) was shown. Seed sequence was highlighted in red in miR-301a-5p. (H) 3' UTR of CDK6, CDK1 or CHEK2 (3' UTR (WT)) as well as 3' UTR (MT) were cloned into a luciferase reporter vector, which were then transfected with control mimic (CTL mimic) or miR-301a-5p mimic into HEK293T cells followed by luciferase activity measurement. Data presented for miR-301a-5p mimic-transfected cells were fold induction over that of control mimic (±s.e.m., ***P < 0.001, NS: non-significant). (I, J) MCF7 cells transfected with control siRNA (si-CTL) or siRNA specifically targeting circPGR (si-circPGR) in the presence or absence of miR-301a-5p inhibitor were subjected to RT-qPCR (I) or immunoblotting (J) analysis to examine the expression of cell cycle genes as indicated. (K, L) MCF7 cells as described in (I) were subjected to cell proliferation assay (K) and FACS analysis (L) (±s.e.m., *P < 0.05). (M) MCF7 cells were infected with lenti-viral control shRNA (sh-CTL) or shRNA specifically targeting circPGR (sh-circPGR) followed by colony formation assay. (N) Quantification of the crystal violet dye as shown in (M) (± s.e.m., ***P < 0.001).