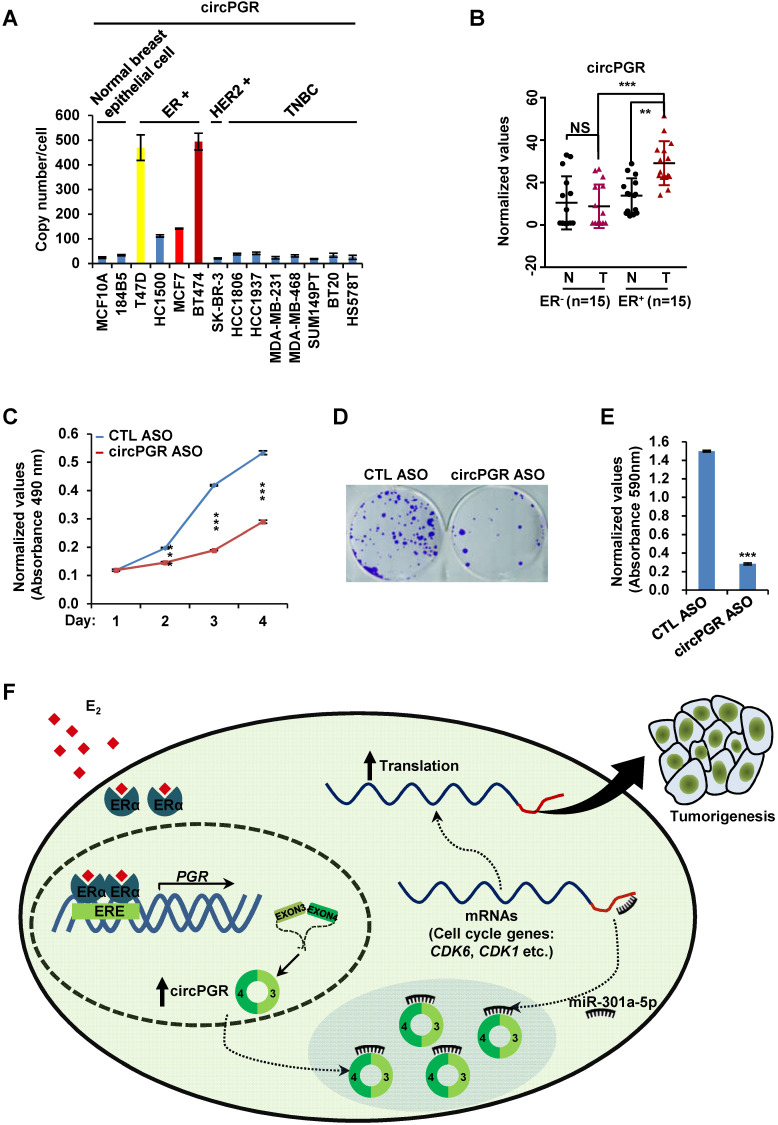
Figure 7.
CircPGR is highly expressed in ER-positive breast cancer cell lines and clinical breast cancer samples, and targeting circPGR is effective in suppressing breast cancer cell growth. (A) RNA samples extracted from 14 different cell lines were subjected to RT-qPCR analysis to examine the expression of circPGR. Data shown was the absolute copy number. Normal breast epithelial cells: MCF10A and 184B5; ER-positive (ER+) cells: T47D, HC1500, MCF7 and BT474; HER2 (human epidermal growth factor receptor 2)-positive (HER2+) cells: SK-BR-3; TNBC (triple-negative breast cancer) cells: HCC1806, HCC1937, MDA-MB-231, MDA-MB-468, SUM149PT, BT20 and HS578T. (B) RNA samples extracted from 15 pairs of ER-negative breast tumor and adjacent normal tissues, and 15 pairs of ER-positive breast tumor and adjacent normal tissues were subjected to RT-qPCR analysis to examine the expression of circPGR. Data shown was normalized to actin (±s.e.m., **P < 0.01, ***P < 0.001, NS: non-significant). (C, D) MCF7 cells transfected with control ASO (CTL ASO) or ASO specifically targeting circPGR (circPGR ASO) were subjected to cell proliferation assay (C) or colony formation assay (D) (±s.e.m., ***P < 0.001). (E) Quantification of the crystal violet dye as shown in (D) (± s.e.m., ***P < 0.001). (F) A proposed model of circPGR function in ER-positive breast cancer: in the presence of estrogen, circPGR was co-induced with its parental gene, PGR, and later stably localized in the cytosol of cells, where it acted as a ceRNA to sponge miR-301a-5p to regulate the expression of multiple cell cycle genes. Alterations in estrogen signaling will result in the aberrant expression of circPGR and the release of cell cycle genes, such as CDK6, CDK1 and CHEK2, from the repression by miR-301a-5p. Aberrant expression of these cell cycle genes will eventually drive breast tumorigenesis.