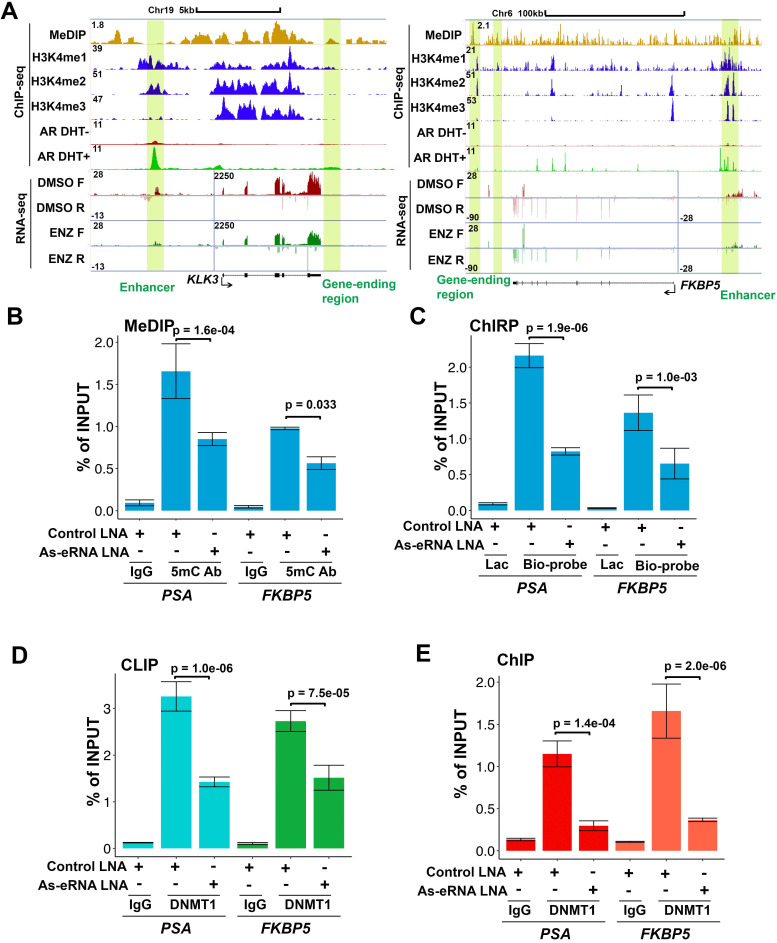
Figure 2.
DNA methylation in antisense promoter is critical for expression of antisense ncRNA. (A) Screen shots from UCSC genome browser showing signal profiles of eRNA and mRNA expression in LNCaP and C4-2. MeDIP-seq in Abl cells are shown as a reference. PSA is in left panel; FKBP5 is in right panel. The enhancer regions are highlighted in yellow box. (B) Quantitative PCR verification of MeDIP results on the antisense enhancer (gene-ending region) of KLK3 and FKBP5 with DNA 5mC antibody against the 5mC sites in C4-2 cells. Means and standard deviations (error bar) were determined from three replicates. Error bars represented mean ± SD for triplicate experiments. P values were shown in the figures. (C) ChIRP assay using biotin-labeled LacZ or antisense eRNA (KLK3 or FKBP5)-specific DNA probes and streptavidin beads. Pull-down DNA was analyzed by real-time PCR. All data shown were mean values ± SD (error bar) from three replicates. P values were shown in the figures. (D) CLIP-qPCR analysis of DNMT1 binding at the KLK3 and FKBP5 antisense RNA in C4-2 cells transfected with control or antisense-specific LNAs. Immunoprecipitated RNAs were detected by real-time PCR. All data shown were mean values ± SD (error bar) from three replicates. P values were shown in the figures. (E) ChIP-qPCR analysis of DNMT1 binding at the KLK3 and FKBP5 gene-ending region in C4-2 cells transfected with control or antisense-specific LNAs. Immunoprecipitated DNAs were detected by real-time PCR. All data shown were mean values ± SD (error bar) from three replicates. P values were shown in the figures.