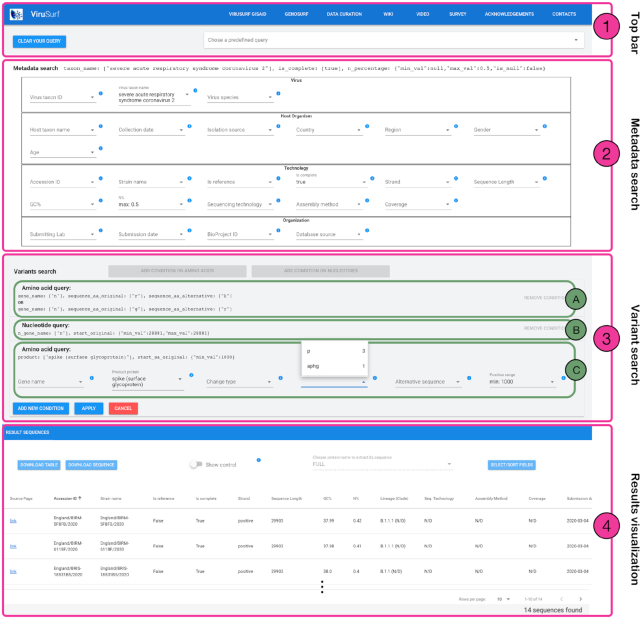
Figure 4.
Overview of ViruSurf interface. Part 1 (Top bar) allows to reset the previously chosen query or select predefined example queries. Queries are composed by using Part 2 (Metadata search) and Part 3 (Variants search). In our example, Part 2 includes three filters on Virus taxon name, Is complete and N%. Part 3 includes three panels. Panel ‘A’ is a query on amino acid variants, selecting sequences with RK and GR changes in gene N; Panel B’ is a query on nucleotide variants, selecting sequences with a variant at position 28 881. Panels ‘A’ and ‘B’ are closed, they can be removed but not changed. Panel ‘C’ is another query on amino acid variants, currently open; it includes two filters selecting given positions of the Spike protein, and visualizes available values for the original amino acid involved in the change. Part 4 shows the Result Visualization. Resulting sequences already reflect the filters of Part 2 and the queries of the closed panels ‘A’ and ‘B’ of Part 3, applied in conjunction. Results can be downloaded, in csv or FASTA format; they can be selected as either cases (default) or controls (switch), and both the nucleotide and amino acid sequences can be projected on a specific protein; table columns can be omitted and reordered. On the bottom right corner, the number of sequences resulting from the search is visualized (in the Figure we show only three sequences out of 14 sequences found).