Figure 5.
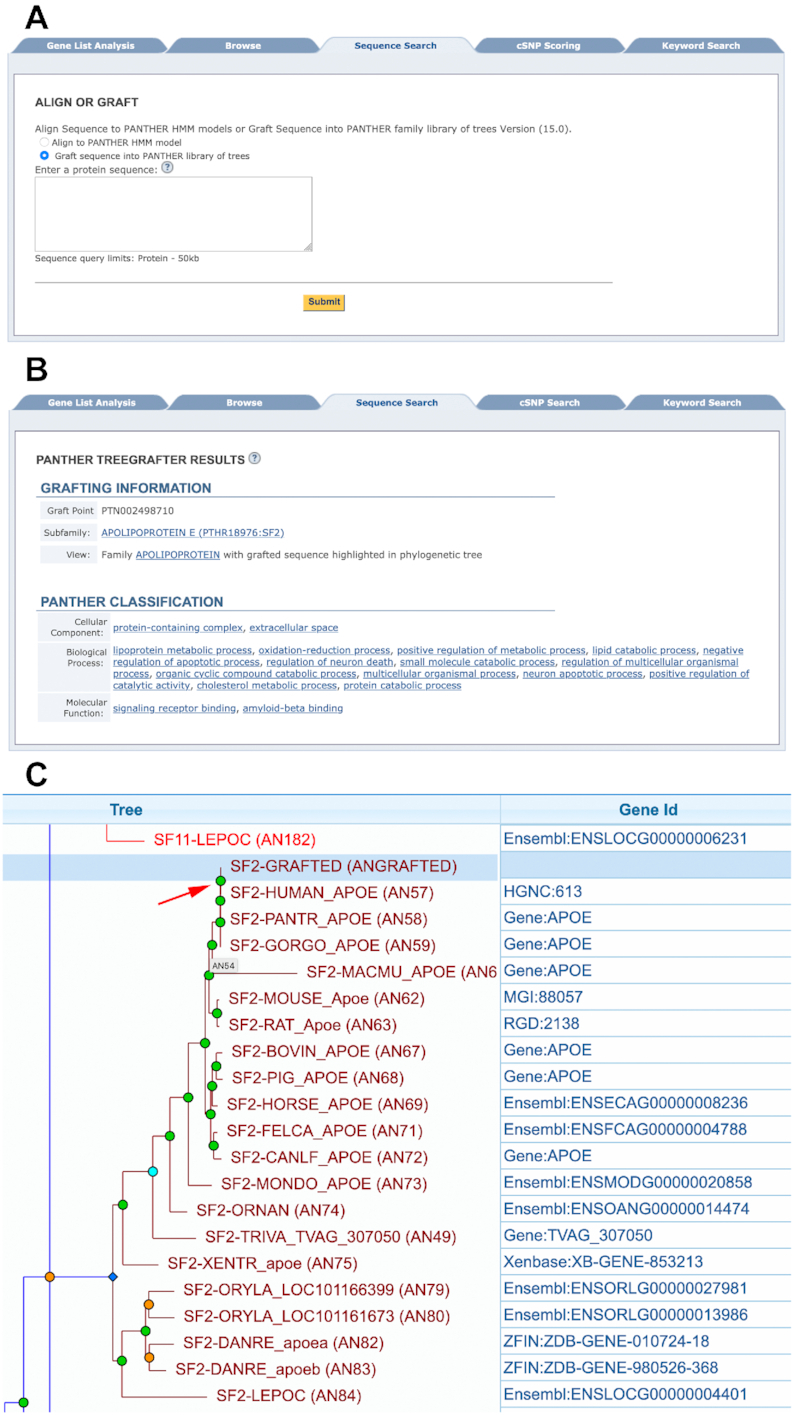
Using TreeGrafter to classify a new protein sequence. (A) Users can classify new sequences using HMMs, or TreeGrafter. (B) TreeGrafter results show subfamily assignment, and GO terms that depend on the graft point in the tree. (C) Users can view the grafted sequence in the context of the reference phylogenetic tree. The new sequence is an additional leaf in the tree (marked ‘ANGRAFTED’ and highlighted in blue), with the new internal node (pointed by a red arrow) induced by the grafting (marked ‘ANINDUCED’).
