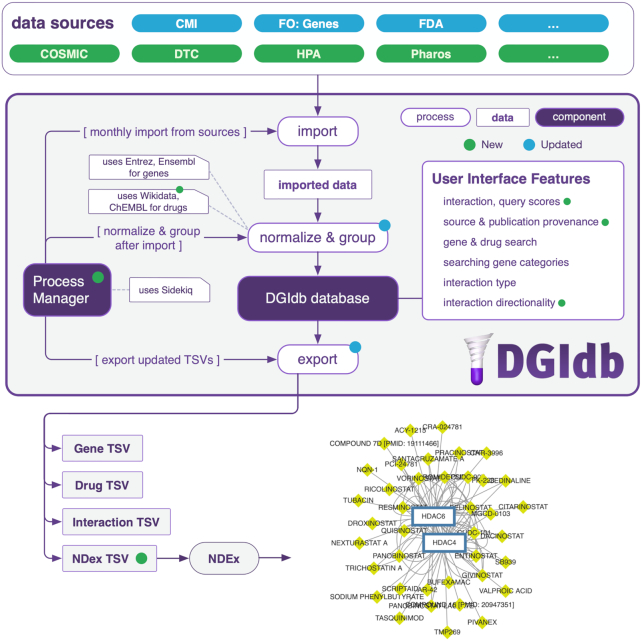
Figure 1.
Overview of main components of DGIdb. Data sources are imported from outside resources (over 40 as of DGIdb 4.0), normalized and grouped with internal processes to prepare records to be displayed in DGIdb, and exported to TSV for download and integration with other resources. Process management is handled by Sidekiq for automation of importing, normalization and grouping, and exporting. A subset of new data sources are highlighted in green, a subset of updated pre-existing data sources are highlighted in blue. The updated sources highlighted in this figure are some of the sources that have been updated through manual curation. Information on additional sources and their status in DGIdb 4.0 can be found in Figure 2 and Supplementary Table S1. New features and technologies from DGIdb 4.0 are indicated with green dots, pre-existing features and technologies that have been updated are indicated with blue dots. The drug-gene network graph shown in the bottom right is an example of the data visualizations available on NDEx. Abbreviations: CMI = Caris Molecular Intelligence, FO = Foundation One, DTC = Drug Target Commons and HPA = Human Protein Atlas.