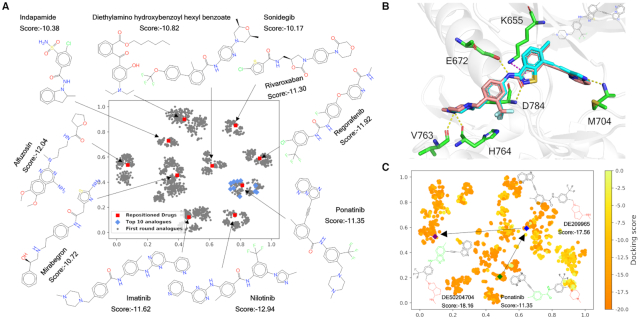
Figure 5.
Docking-based virtual screening of DDR1 inhibitors against DrugSpaceX compounds. (A) The t-SNE projection of the compounds in Set1, including the top 10 drugs repositioned in relation to DDR1 and their first round of transformation products. The chemical structure features were encoded as an ECFP4 512-bit vector for t-SNE analysis. (B) The putative binding mode of DE209841 (cyan carbons) derived from docking simulations compared to the crystallized ligand ponatinib (salmon carbons) in DDR1 kinase (PDB code: 3ZOS). (C) The t-SNE projection of the compounds in Set2 coloured by docking scores ranging from the lowest in orange to the highest in yellow. The compound DE50204704, showing the lowest docking score, can be traced back to ponatinib in two rounds of transformation.