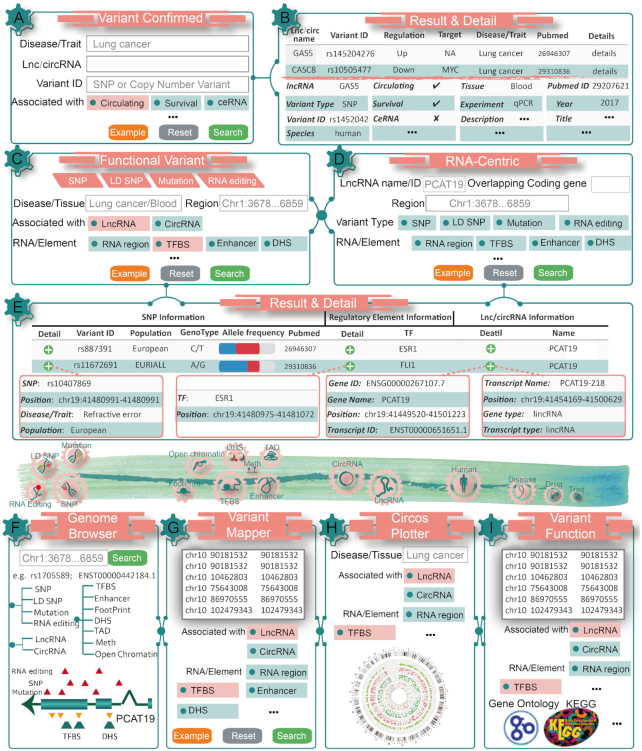
Figure 2.
Workflow and examples of using LincSNP 3.0. (A) The variant confirmed search interface of LincSNP 3.0 by querying an example of lung cancer. (B) The query results and detailed information of variants, RNAs, disease, biomarkers and literatures in LincSNP 3.0. (C) The functional variant search interface of LincSNP 3.0 by querying an example of lung cancer and blood. (D) The RNA-centric search interface of LincSNP 3.0 by querying an example of lncRNA PCAT19. (E) The query results and detailed information of variants, RNAs and regulatory elements in LincSNP 3.0. (F) The variants and regulatory elements landscape of PCAT19 in LincSNP 3.0 Genome Browser. (G) Interested variants map to lncRNAs and circRNAs in LincSNP 3.0 Variant Mapper. (H) The circos plot diagram including variants, regulatory and lncRNAs for lung cancer in LincSNP 3.0 Circos Plotter. (I) The functional enrichment analysis for variants in LincSNP 3.0 Functional Annotation.