Figure 1.
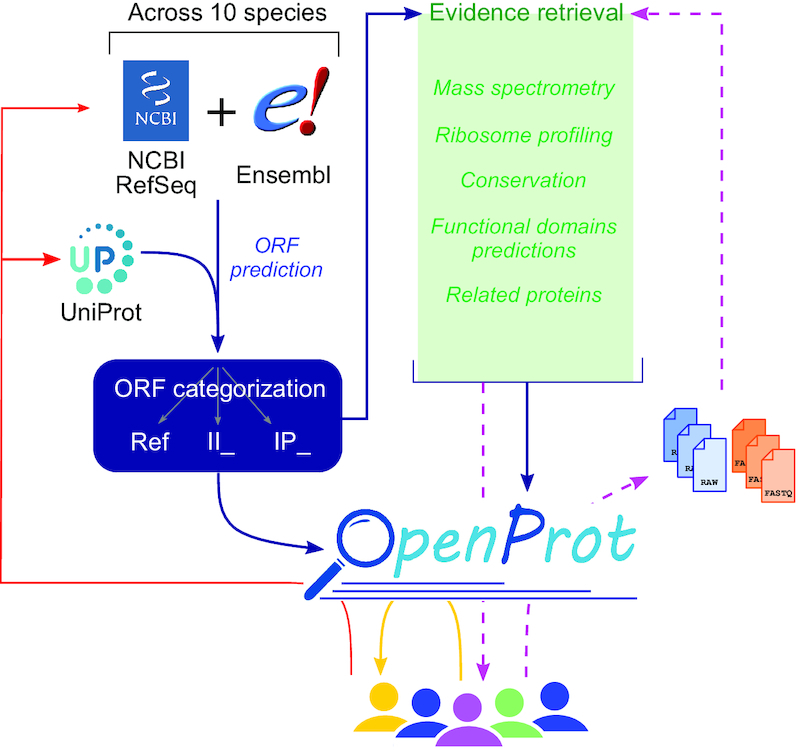
The OpenProt (v1.6) proteogenomic resource. OpenProt pipeline (dark blue arrows) contains two main features: prediction (on the left, blue) and evidence retrieval (middle, green). OpenProt enforces a polycistronic model of eukaryotic genes contrary to the actual dogma of one CDS per transcript. It retrieves all possible ORFs from transcripts annotated in NCBI RefSeq and/or Ensembl (ORF prediction, blue). The ORF-encoded proteins are then categorized as follows: RefProt if already annotated in NCBI RefSeq, Ensembl and/or UniProt; novel isoforms of known CDS (II_ accessions); or novel alternative proteins (IP_ accessions). All predicted proteins are available on the OpenProt website (www.openprot.org). Furthermore, OpenProt retrieves supporting evidence for all proteins from MS, ribosome profiling, protein conservation, functional domain prediction and related proteins. All data are inserted in the OpenProt resource and freely available to the community (yellow arrow). OpenProt also allows data submission by users for analysis using the OpenProt pipeline (purple dashed arrows). The results are then returned to the users and inserted in OpenProt at the next release (purple dashed arrows). With a symbiotic behaviour between the scientific community, linking experimental data, bioinformatics resources and deep ORF annotation, OpenProt participates in the implementation of novel, experimentally supported proteins in genome annotations and protein databases (red arrows).
