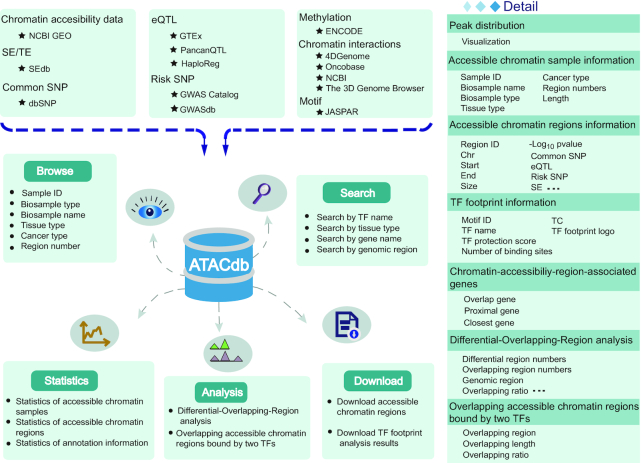
Figure 1.
Database content and construction. Chromatin accessibility regions in ATACdb were calculated based on human ATAC-seq data. Genetic and epigenetic annotations were collected or calculated, including super-enhancers, typical enhancers, TFs, common SNPs, risk SNPs, eQTLs, LD SNPs, DNA methylation sites, 3D chromatin interactions and TADs. Users can determine the scope of the chromatin accessibility data query through four paths: genomic region-based query, tissue-category-based query, TF-based query and gene-based query. ATACdb contains analytical tools and multiple functions to browse, search, download and visualize chromatin accessibility information.