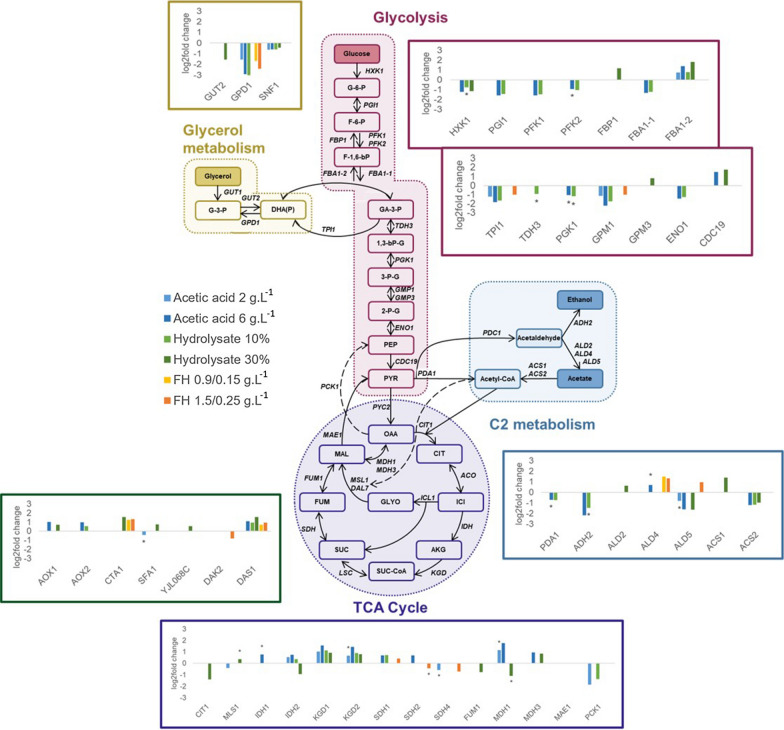
Fig. 4.
Gene expression of central carbon metabolism pathways for P. pastoris. Possible carbon sources are presented in colored squares: glucose; glycerol, ethanol, acetate. Bar charts represent the transcriptional changes (log2 fold) of genes in acetic acid 2 g.L−1 (light blue), 6 g.L−1 (dark blue), FH 0.9/0.15 g.L−1 (yellow), FH 1.5/0.25 g.L−1 (orange), hydrolysate 10% (light green) and 30% (dark green) with P-values < 0.05 or 0.1 (*on top of bar chart). Metabolites: G-6-P glucose 6-phosphate, F-6-P fructose-6-phosphate, F-1,6-P fructose 1,6-phosphate, G-3-P glycerol 3-phosphate, GA-3-P glyceraldehyde 3-phopshate, 1,3-bPG 1,3-bisphosphoglycerate, 3-PG 3-phosphoglycerate, 2-PG 2-phosphoglycerate, PEP phosphoenolpyruvate, PYR pyruvate, DHA(P) dihydroxy acetone (phosphate), OAA oxaloacetate, CIT citrate, ICI isocitrate, AKG alpha-keto glutarate, SUC succinate, SUC-CoA succinyl-Coenzyme A, FUM fumarate, MAL malate, GLYO glyoxylate. Enzymes: HXK1 hexokinase, PGI1 phosphoglucose isomerase, PFK1/2 phosphofructokinase, FBP1 fructose-1,6-bisphosphatase, FBA1-1/1-2 fructose 1,6-bisphosphate aldolase, TPI1 triose phosphate isomerase, TDH3 glyceraldehyde-3-phosphate dehydrogenase, PGK1 3-phosphoglycerate kinase, GPM1/3 phosphoglycerate mutase, ENO1 enolase I, phosphopyruvate hydratase, CDC19 pyruvate kinase, GUT1 glycerolkinase, GUT2 glycerol-3-phosphate dehydrogenase, GPD1 glycerol-3-phosphate dehydrogenase, SNF1 central kinase, PYC2 pyruvate carboxylase, CIT1 citrate synthase, ACO1/2 aconitase, ICL1 isocitrate lyase, DAL7 malate synthase, IDH1/2 isocitrate dehydrogenase, KGD1 alpha-ketoglutarate dehydrogenase complex, KGD2 dihydrolipoyl transsuccinylase, LSC1 succinyl-CoA ligase, SDH1/2/4 succinate dehydrogenase, FUM1 fumarase, MDH1 mitochondrial malate dehydrogenase, MDH3 malate dehydrogenase, MAE1 mitochondrial malic enzyme, PDC1 pyruvate decarboxylase, PDA1 pyruvate dehydrogenase (subunit from PDH complex), ALD2 cytoplasmic aldehyde dehydrogenase, ALD4-1/4-2/5 mitochondrial aldehyde dehydrogenase, ADH2 alcohol dehydrogenase, ACS1/2 acetyl-coA synthetase, PCK1 phosphoenolpyruvate carboxykinase. Genes or conditions with P-values out of the threshold were not depicted