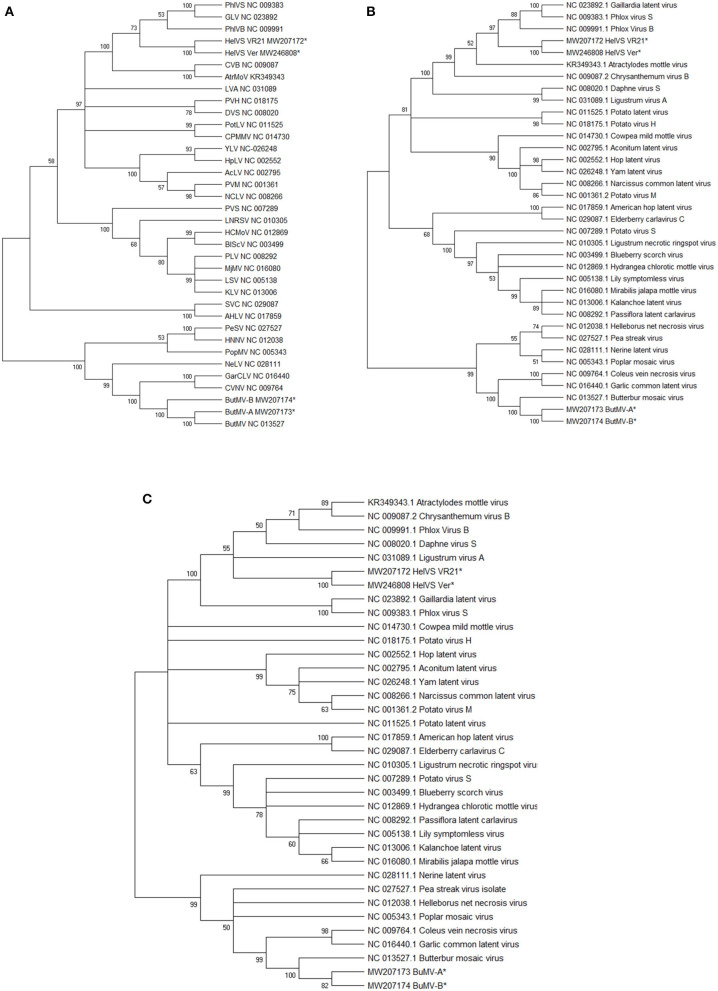
Figure 5.
(A–C) Maximum Likelihood phylogenetic trees with 1,000 bootstrap replications of: (A), the genomic nucleotide sequences; (B) the Replicase (RdRp) amino acid sequences; and (C) the Coat protein (CP) amino acid sequences. Branches with <50% bootstrap support were collapsed. The GenBank accession number for the genomes are listed following the virus acronym in (A); the RdRp and CP sequences were taken from the genome accessions. Virus abbreviations: AcLV, Aconitum latent virus; AHLV, American hop latent virus; AtrMoV, Atractylodes mottle virus; BlScV, blueberry scorch virus; ButMV, butterbur mosaic virus; CPMMV, cowpea mild mottle virus; CVB, chrysanthemum virus B; CVNV, coleus vein necrosis virus; DVS, Daphne virus S; GarCLV, garlic common latent virus; GLV, Gaillardia latent virus; HCMoV, hydrangea chlorotic mottle virus; HelVS, Helenium virus S; HNNV, Helleborus net necrosis virus; HpLV, hop latent virus; KLV, kalanchoe latent virus; LNRSV, Ligustrum necrotic ringspot virus; LSV, lily symptomless virus; LVA, Ligustrum virus A; MjMV, Mirabilis jalapa mottle virus; NCLV, narcissus common latent virus; NeLV, Nerine latent virus; PeSV, pea streak virus; PhlVB, phlox virus B; PhlVS, phlox virus S; PLV, Passiflora latent virus; PopMV, poplar mosaic virus; PotLV, potato latent virus; PVH, potato virus H; PVM, potato virus M; PVS, potato virus S; SVC, Sambucus virus C; YLV, Yam latent virus. The next-generation sequences of both isolates of HelVS, and both isolates of ButMV from the current studies are included in each tree, and each is marked with an asterisk (*).