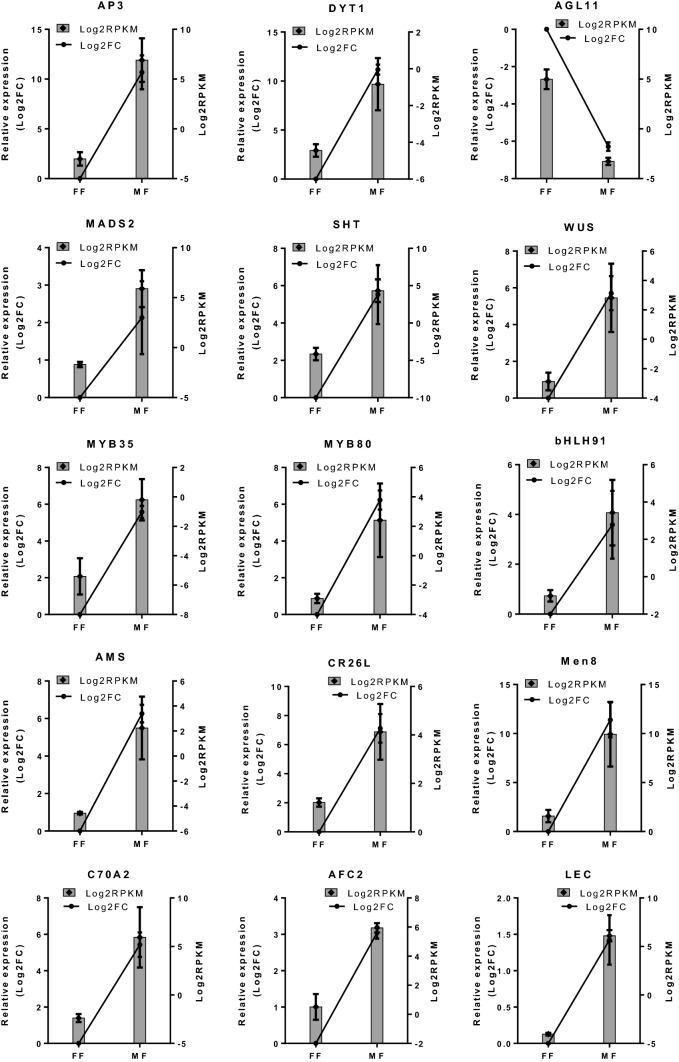
Fig. 6.
qPCR assays of selected DEGs in flowers of normal male (MF) and female (FF) plants. These DEGs include nine transcription factor genes: APETALA 3 (AP3), Dysfunctional Tapetum1 (DYT1), Agamous-like MADS-box 11 (AGL11), MADS2, WUSCHEL (WUS), MYB35, MYB80, bHLH91and ABORTED MICROSPORES (AMS), and six other genes: Spermidine hydroxycinnamoyl transferase (SHT), Eceriferum 26-like (CR26), MEN-8 (male-specific protein—Men8), Cytochrome P450 703A2 (C70A2), Serine threonine- kinase AFC2 (AFC2) and Mannose glucose-specific lectin (LEC) involved in floral development and sex determination. qPCR data were expressed as mean values ± standard errors (n = 2) of log2 fold change. The relative expression (qPCR) in the samples of FF was set arbitrary to 1 (log2(1) = 0). RNA-Seq data were shown as mean values ± standard errors (n = 2–4) of log2 RPKM. All of the targeted genes had similar expression patterns in both qPCR and RNA-Seq analyses between the two flower sex types