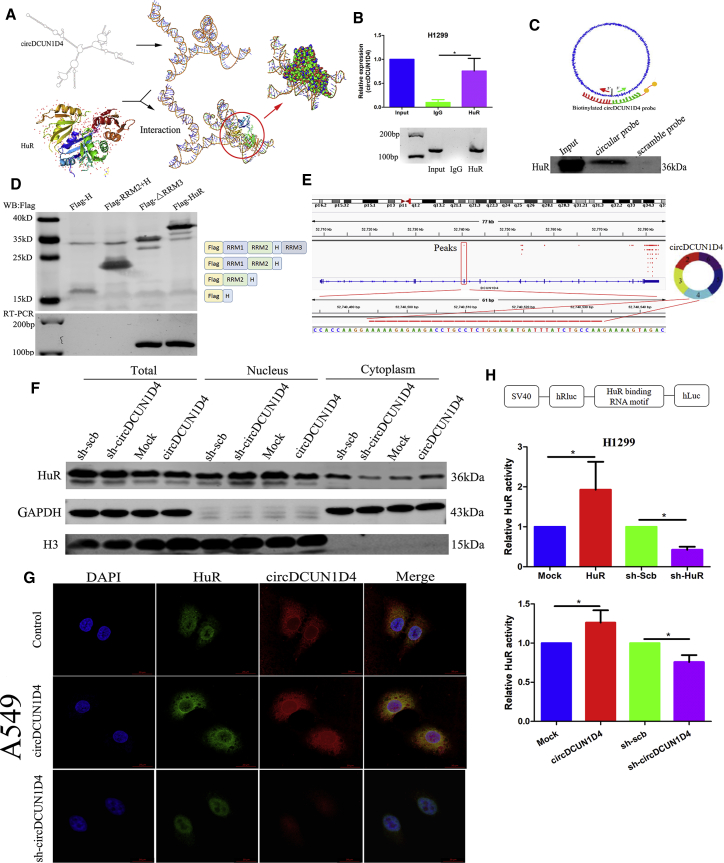
Figure 4.
circDCUN1D4 interacts with and activates HuR protein in cancer cells
(A) Graphical representation of three-dimensional structures of circDCUN1D4 and HuR docking models with a zoom-in image of the binding interface generated by NPDock. (B) RIP assay showing that the RNA binding protein HuR interacts with circDCUN1D4. (C) RNA pull-down assay showing the HuR protein pulled down by circRNA probes or scramble probes from lysates of H1299 cells. (D) RIP assay depicting the recovered circDCUN1D4 levels from H1299 cells detected by RT-PCR (lower panel) after incubation with full-length or truncated forms of Flag-tagged recombinant HuR protein validated by western blot (upper panel). (E) IGV showing the peaks localized in exon 4 of circDCUN1D4, which interacts with the HuR protein. (F) Western blot indicating the expression of HuR in total lysates or subcellular fractions of H1299 cells stably transfected with mock, circDCUN1D4, sh-scb, or sh-circDCUN1D4 vectors. (G) Dual RNA-FISH and immunofluorescence staining assay showing the colocalization of circDCUN1D4 (red) and HuR (green) and the translocation of HuR (green) from the nucleus to the cytoplasm with DAPI (blue). Scale bar, 5 μm. (H) Dual-luciferase assay (middle and lower panels) indicating the activity of the HuR reporter (upper panel) in H1299 cells stably transfected with mock, HuR, circDCUN1D4, sh-scb, sh-HuR, or sh-circDCUN1D4 vectors (mean ± SD, n = 4). Student’s t test compared the difference in (H). ∗p < 0.05 versus mock or sh-scb.