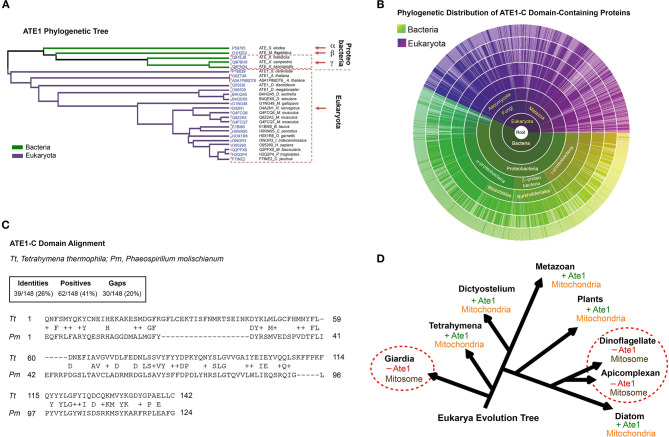
Figure 1.
The alpha-proteobacterial origin of ATE1 links the protein to mitochondria. (A) Cluster analysis of ATE1 proteins in evolutionary diverse organisms, using the Clustal Omega program on the Uniprot website. The phylogenetic tree is presented as a Notug 2.6 graph. It highlights the clustering of ATE1 from alpha-, beta-, and gamma-proteobacteria, as well as eukaryota. (B) Sunburst graph showing the distribution of the catalytic core ATE1-C domain (Pfam ID: PF04377), among 1,796 different species currently known to contain such a sequence encoded in their genomes. The graph was generated with tools from pfam.xfam.org hosted by EMBL-EBI. Yellow-green colors represent different types of bacteria, and purple color represent eukaryotes. No entry from archaea was found in this database. In addition, manual searches for ATE1 homologs on the genome of a representative archaea (Haloferax volcanii) using the BlastP function from the NCBI database and the ATE1 amino acid sequence from yeast, mouse, or alpha-proteobacteria as inquiries returned no significant homologous hits. (C) Comparison of the amino acid sequences of the core domain of ATE1 (ATE-C domain; Pfam ID: PF04377) between a eukaryote (Tetrahymena thermophila, strain SB210) and a alpha-proteobacterium (Phaeospirillum molischianum DSM). The sequence alignment was performed with NCBI BLASTp. (D) Illustration of the eukarya evolution tree, showing the relationship between the presence of an ATE1 gene and the mitochondrial development state in several eukaryotic species. The red circles highlight several families (giardia, dinoflagellate, and apicomplexan) in which the absence of ATE1 is accompanied by a loss of respiratory function in mitochondria, organelles that are reduced to a minimized form known as mitosomes.