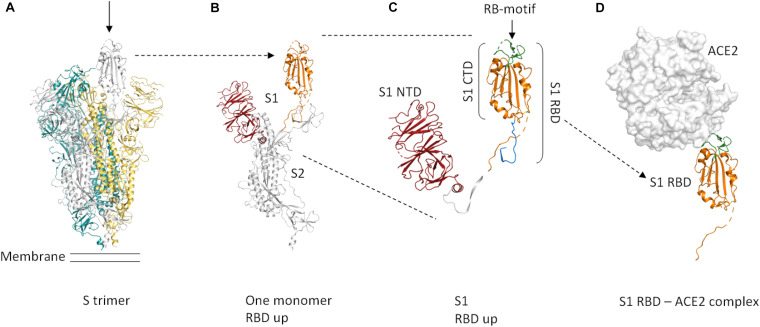
FIGURE 1.
Cartoon representation of the protein domains of the 2019-nCoV spike protein. Dashed lines in the structure indicate flexible regions that are not defined. Glycosylations are not shown. The domain boundaries are based on UniProt entry P0DTC2. (A) The S protein forms a trimer that is exposed on the virus surface. The monomers forming the trimer are shown in gray, yellow, and teal. In the prefusion conformation shown here (PDB entry 6VSB, Wrapp et al., 2020), only one of the three RBD domains is rotated up (chain shown in gray, arrow) in a conformation that is accessible for ACE2 binding. (B) Only the chain shown in gray in (A) is depicted here. A selection of important features is shown in color (orange = S RBD amino acids 319–541, red = S1 NTD. (C) Close-up view of S1, showing the S1 NTD (amino acids 13–303, red), S1 CTD (amino acids 334–527, orange and green), S_RBD as in our construct (amino acids 319–527, shown in orange and green) and the receptor-binding motif (amino acids 437–508, green). The linker that connects S RBD and S1 NTD is shown in white. The stretch shown in blue comprises amino acids 528–541. These most C-terminal residues of S RBD are not included in our construct. (D) ACE2 binding to S RBD. The ACE2 peptidase domain is shown as a surface representation (white). S RBD is shown in orange and green. The green region indicates the receptor-binding motif. For this Figure, the S RBD of PDB entry 6M17 (Yan et al., 2020) was superimposed onto the S RBD of PDB entry 6VSB (Wrapp et al., 2020). The S RBD of PDB entry 6VSB and the ACE2 peptidase domain of PDB entry 6M17 are shown. The Figure was created using PyMOL (The PyMOL Molecular Graphics System, Version 2.3.4 Schrödinger, LLC).