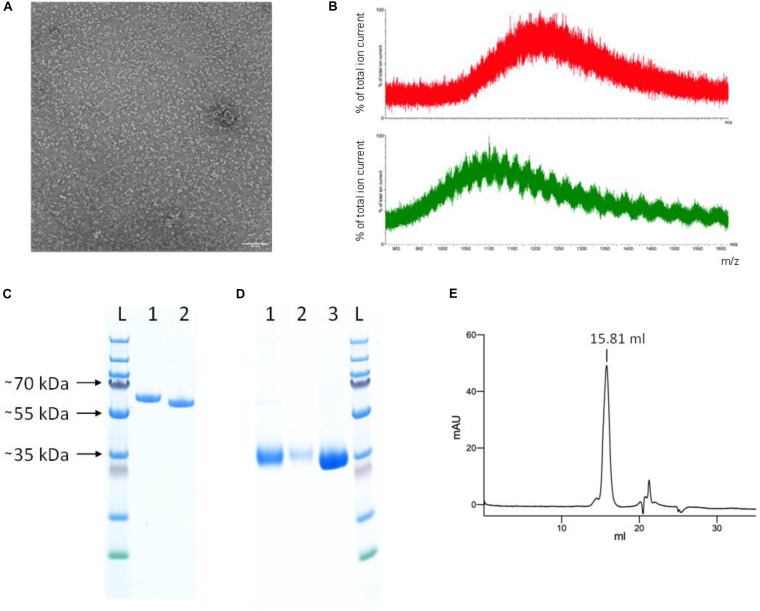
FIGURE 4.
Protein characterization. (A) Negative staining electron micrograph of SEC-purified YFP-S_RBD fusion protein Scale bar 50 nm. (B) Mass spectrometry results of untreated (red, top) and PNGase F treated (green, bottom) YFP-S_RBD fusion protein. The data shows that the protein contains glycosylations. Due to glycosylation heterogeneities, a clear mass could not be determined. PNGase F treatment reduced the mass but did not remove all glycosylations, indicating that PNGase F—resistant glycosylations, such as O-linked glycosylations, are also present. (C,D) SDS-PAGE analysis. (C) YFP-S_RBD. L, Ladder; 1, Reducing SDS sample buffer; 2, Non-reducing sample buffer. (D) PreScission-cleaved YFP-S_RBD. 1, S_RBD from Ni-NTA IMAC; 2, S_RBD from SEC; 3, YFP washed off the Ni-NTA resin after on-bead cleavage, after removal of the protease. (E) Analytical SEC profile (280 nm absorbance) of S_RBD. Based on a calibration curve using the bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa) and horse myoglobin (17 kDa) peaks, the retention volume suggests an approximate molecular mass of 37 kDa. The calculated molecular weight of S_RBD, based on protein sequence, excluding glycosylations, is 25 kDa. The SDS-PAGE (shown in D) indicates a size of ∼35 kDa for the glycosylated S_RBD.