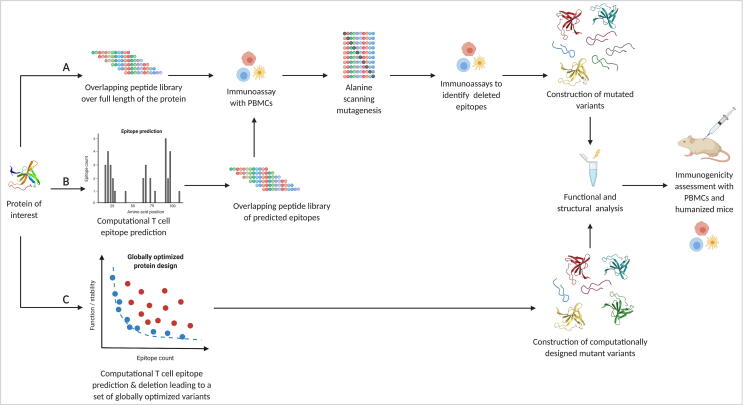
Fig. 2.
Overview of T cell epitope deletion workflows. T cell epitope deletion transitioned from being purely experimental to more and more computational. A) This pathway illustrates the steps for experimental T cell epitope deletion. The most common way was to synthesize an overlapping peptide library over the full length of the protein of interest. Epitopes among those peptides are identified through ex vivo immunoassays with peripheral blood mononuclear cells (PBMCs) from a broad range of healthy donors. The immunogenic peptides are subjected to alanine scanning mutagenesis and then tested again for immunogenicity with the PBMCs. Confirmed deimmunizing mutations can then be combined and introduced in the full length protein. Introducing mutations comes with a high risk of misfolding and inactivation of the protein. Therefore, functional and structural characterisation is needed. Deimmunization success of a functional protein can be tested and validated in humanized mice and with PBMCs. B) The use of epitope prediction reduces the number of peptides to be tested for immunogenicity by only testing overlapping peptides in predicted epitope regions. The subsequent steps are identical to the experimental strategy. C) The computational approach combines epitope prediction with epitope deletion in one program by introducing mutations in silico while taking into account the global effect of those changes. Independent of the strategy chosen, deimmunized variants have to be produced and tested experimentally. (Created with BioRender.com).