Figure EV3. 2D gel electrophoresis of the DNA linking number distribution of circular minichromosomes.
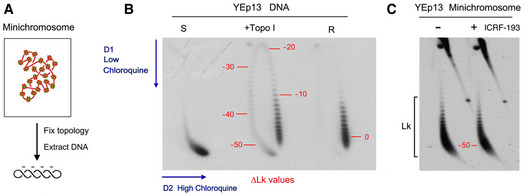
- DNA molecules extracted from yeast circular minichromosomes are negatively supercoiled since each nucleosome constrains about one negative supercoil (∆Lk ≈ −1).
- 2D gel electrophoresis of covalently closed DNA circles, in which the first and second gel dimensions are run in the presence of low and high concentrations of chloroquine, respectively, allow the Lk distribution of DNA topoisomers to be resolved along an arch, in which Lk values increase clockwise. The 2D gel shows highly negatively supercoiled (S), partially relaxed (+topo I), and fully relaxed (R) forms of the YEp13 plasmid. Numbers in red indicate approximate ∆Lk values relative to the center of the relaxed (R) Lk distribution (∆Lk = 0).
- 2D gel electrophoresis of DNA of the YEp13 minichromosome (as in Fig 2C). Comparison of the gels in B and C indicates that DNA in the YEp13 minichromosome is negatively supercoiled and has ∆Lk close to −50. This value is consistent with the plausible number of nucleosomes assembled in YEp13 (10.7 Kb). However, note that since the outline of Lk distributions can vary in separate 2D gels (i.e., due to differences in tank dimensions, power supply and temperature during electrophoresis), only DNA samples that ran in the same gel (side by side) can be accurately compared.
