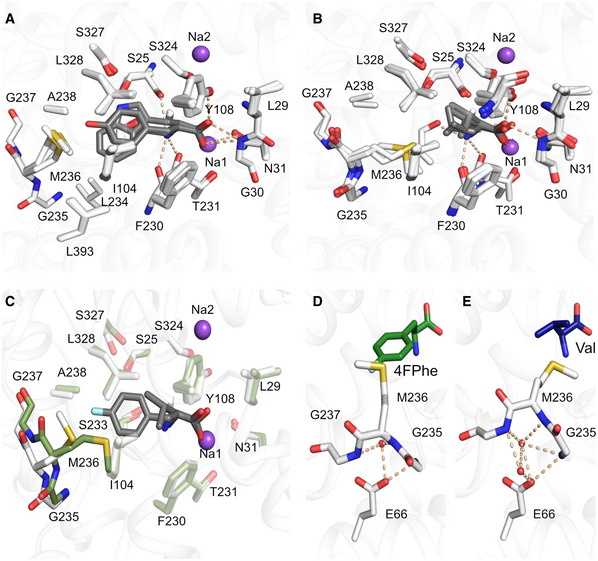
Figure 3. Binding of aliphatic and aromatic substrates to MhsT.
-
A, BGrouping of binding sites: (A) aromatic ligands consisting of L‐Trp, L‐Phe, L‐4‐F‐Phe, and L‐Tyr, (B) aliphatic ligands consisting of L‐Val, L‐Leu, and L‐Ile.
-
COverlay of MhsT‐Val (in green) and MhsT‐4FPhe (in white) structures to visualize changes upon binding of different sized ligands. The unwound region of TM6 is non‐transparent.
-
D, EConformation of the unwound part of TM6 in case of an aromatic substrate (L‐4‐F‐Phe as an example) and (E) aliphatic substrate (L‐Val as an example). Glu66 is coordinating the GMG loop through water molecules—in the case of aromatic substrates only one water molecule is found, whereas with aliphatic substrates two water molecules are present.
Data information: Protein is shown as white ribbon with the binding site defined with white sticks. Substrates are visualized as gray sticks and the sodium ions as purple spheres.
