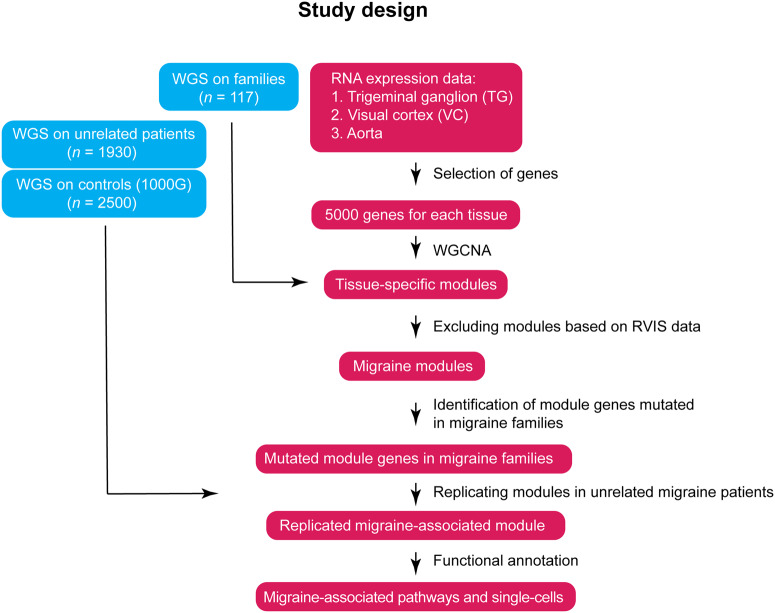
Figure 1.
Study design. Co-expression networks were constructed for the trigeminal ganglion, visual cortex and aorta, to identify tissue-specific modules of highly co-expressed genes. In parallel, the WGS data were analysed to identify mutations. Modules with over-representations of mutations were defined as potential ‘migraine modules’. Similarly, we defined mutations in an independent cohort compared to population controls and replicated the potential migraine modules. Replicated modules were further analysed using functional annotation. RVIS = residual variation intolerance score.