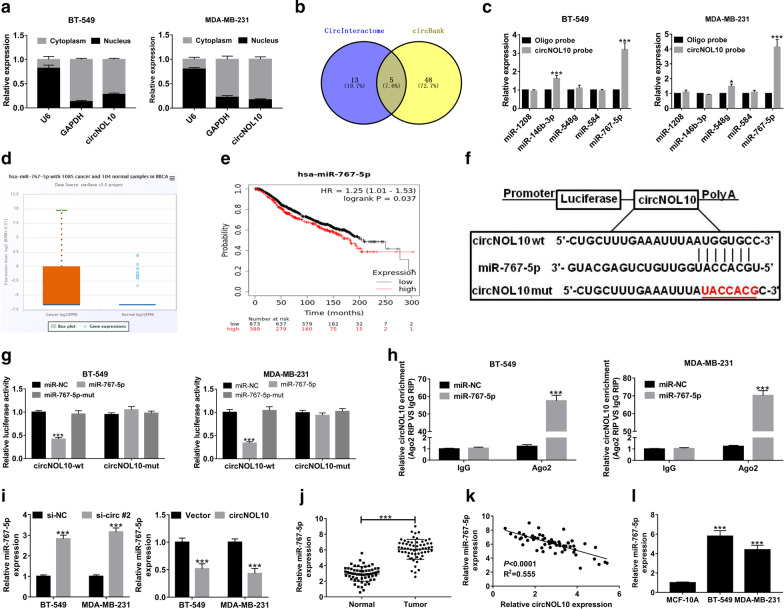
Fig. 4.
CircNOL10 acts as a sponge for miR-767-5p in BC cells. a Subcellular localization analysis was conducted to measure the percentage of circNOL10 expression in nucleus and cytoplasm of BC cells. b The candidate miRNAs containing the putative binding sites of circBOL10 were predicted via CircInteractome and CircBank online databases. c RNA pull down experiments were performed by using biotinylated circNOL10 probe, followed by qRT-PCR analysis of the candidate 5 miRNAs. d Expression level of miR-767-5p was analyzed in 1085 tumor tissues and 104 normal samples in breast invasive carcinoma (BRCA) by starBase Pan-Cancer Analysis Platform. e Kaplan–Meier survival analysis of TCGA cohort to examine the correlation between the overall survival and miR-767-5p expression in BC patients. f The predicted binding sequences between circNOL10 and miR-767-5p by web-based softwares. The red section is a sign of mutated bases. g The luciferase activities of circNOL10-wt and circNOL10-mut reporters in BT-549 and MDA-MB-231 cells transfected with miR-NC, miR-767-5p or miR-767-5p-mut were measured by dual-luciferase reporter assays. h RIP assays were conducted by using Ago2 or IgG antibody in BT-549 and MDA-MB-231 cells transfected with miR-NC or miR-767-5p, followed by the detection of circNOL10 enrichment in immunoprecipitates. i qRT-PCR was used to examine the level of miR-767-5p in BT-549 and MDA-MB-231 cells with circNOL10 knockdown or overexpression. j The level of miR-767-5p in 60 pairs of BC tissues and adjacent normal tissues. k Pearson’s correlation coefficients were used to assess the correlation between circNOL10 and miR-767-5p in BC tissues (n = 60). l Expression difference of miR-767-5p in BC and MCF-10A cells. ***P < 0.001