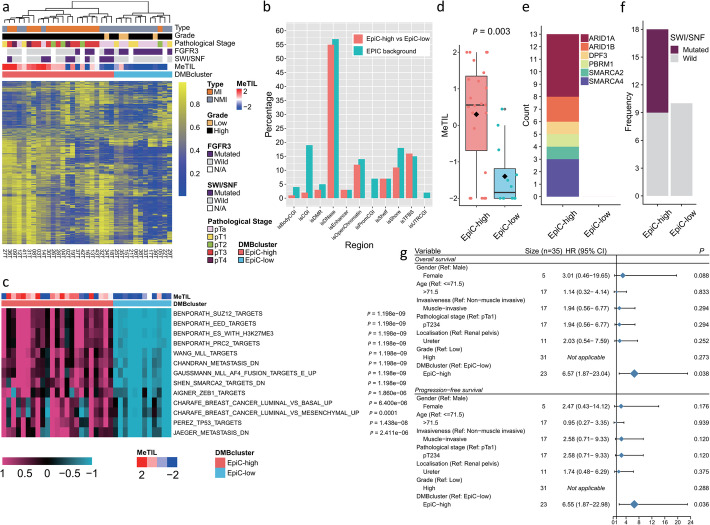
Fig. 5.
Analysis of upper tract urothelial carcinomas (UTUC) DNA methylation. a Unsupervised clustering of most variable DNA methylation probes in UTUC showing two epi-clusters: EpiC-C1(high) and EpiC-C2(low). b Percentage and distribution of differentially DNA methylation probes (blue) between EpiC-high and EpiC-low subgroups across different genomic regions annotated by EPIC arrays (red). Note that there is no enrichment identified in specific genomic regions. c Single-sample Gene Set Enrichment Analysis (ssGSEA) showing pathways enriched in EpiC-high relative to EpiC-low cluster. d Boxplot showing significantly higher MeTIL score in EpiC-high subgroup than that in EpiC-low subgroup (P = 0.003). e Count of mutations in SWI/SNF family genes in EpiC-high and EpiC-low clusters. f Frequency of mutations affecting SWI/SNF family genes in EpiC-high versus EpiC-low UTUC subgroups. Note that 9 out of 18 (50%) UTUC samples in the EpiC-high subgroup harbored mutations as compared to none in the EpiC-low subgroup. g Forest plot showing that the DMBcluster of EpiC-high and EpiC-low is a significant prognostic factor as compared to other major clinicopathological features for either overall survival or progression-free survival