Abstract
Background
Glioblastoma (GBM) is the most common primary malignant brain tumor in adulthood. Despite multimodality treatments, including maximal safe resection followed by irradiation and chemotherapy, the median overall survival times range from 14 to 16 months. However, a small subset of GBM patients live beyond 5 years and are thus considered long-term survivors.
Methods
A retrospective analysis of the clinical, radiographic, and molecular features of patients with newly diagnosed primary GBM who underwent treatment at The University of Texas MD Anderson Cancer Center was conducted. Eighty patients had sufficient quantity and quality of tissue available for next-generation sequencing and immunohistochemical analysis. Factors associated with survival time were identified using proportional odds ordinal regression. We constructed a survival-predictive nomogram using a forward stepwise model that we subsequently validated using The Cancer Genome Atlas.
Results
Univariate analysis revealed 3 pivotal genetic alterations associated with GBM survival: both high tumor mutational burden (P = .0055) and PTEN mutations (P = .0235) negatively impacted survival, whereas IDH1 mutations positively impacted survival (P < .0001). Clinical factors significantly associated with GBM survival included age (P < .0001), preoperative Karnofsky Performance Scale score (P = .0001), sex (P = .0164), and clinical trial participation (P < .0001). Higher preoperative T1-enhancing volume (P = .0497) was associated with shorter survival. The ratio of TI-enhancing to nonenhancing disease (T1/T2 ratio) also significantly impacted survival (P = .0022).
Conclusions
Our newly devised long-term survival-predictive nomogram based on clinical and genomic data can be used to advise patients regarding their potential outcomes and account for confounding factors in nonrandomized clinical trials.
Keywords: glioblastoma, long-term survival, nomogram, outcome, prediction
Key Points.
Our new long-term survival-predictive nomogram can advise GBM patients on outcomes.
The nomogram may help account for factors confounding nonrandomized clinical trials.
Importance of the Study.
Glioblastoma (GBM) survival is approximately a year. A small subset of GBM patients is afforded long-term survival but the features that predict this outcome are not fully delineated. This study integrates clinical, radiographic, and genomic data to assess the probability of long-term survival. We performed extensive molecular profiling on a cohort of patients with de novo GBM and integrated this data with clinical and radiographic features. We then constructed an integrated predictive outcome nomogram based on age, Karnofsky Performance Scale score, and the mutational status of IDH1, PTEN, and TP53 which was validated with The Cancer Genome Atlas. This long-term survival-predictive score can provide clinicians with a framework to inform patients about their relative chances of long-term survival. The nomogram will also be a valuable assessment tool for clinical trial investigators, regulatory agencies, and other stakeholders to identify confounding variables influencing the interpretation of outcomes data.
The current standard-of-care for treatment of glioblastoma (GBM) is maximal safe resection followed by concurrent chemoradiation and cyclic administration of temozolomide. Despite aggressive, multimodality therapy, the median survival times range from 14 to 16 months.1,2 However, a small percentage of GBM patients (<3%) live well beyond 5 years, suggesting that these long-term survivors have a disease course that is inherently different from that in most GBM patients.3 The factors that define this subset of GBM patients who defy the odds remain largely unknown. Several small series have focused on clinical predictors of outcome,4,5 whereas others have focused on molecular determinants of survival.6–8 Currently available calculators of predictive outcomes in GBM patients are mostly reliant on clinical features (https://www.eortc.be/tools/gbmcalculator/; http://cancer4.case.edu/rCalculator/rCalculator.html). An integrated clinical and molecular framework for long-term survival (LTS) has yet to be devised. To that end, the objective of the present study was to perform an in-depth analysis of a cohort of GBM patients to determine not only the clinical factors but also the molecular determinants of survival. As a result, we developed a survival-predictive nomogram to identify confounders that may be relevant for the assessment of nonrandomized GBM clinical trials and to enable oncologists to more appropriately advise patients regarding their potential outcomes.
Methods
Study Population
This study was conducted under The University of Texas MD Anderson Cancer Center Institutional Review Board-approved protocol PA15-0636 and consisted of 80 patients with newly diagnosed GBM who underwent treatment at MD Anderson from November 1, 1996 to December 31, 2015 and had sufficient quality and quantity of tissue available for next-generation sequencing and immunohistochemical analysis along with full radiographic and clinical annotation. This study strictly excluded secondary IDH-mutant GBM with a multi-year prior history of known low-grade astrocytoma and was focused on de novo, classic spontaneously arising GBM. Patients were stratified into 3 analysis groups: short-term survival (STS; <6 months [n = 37]), median survival (MTS; ~15 months [n = 22]), and LTS (>5 years [n = 21]). Four of the patients in the STS group and 3 of those in the MTS group were censored because they were lost to follow-up (ie, their exact dates of death were unavailable). Tumor diagnosis was made using magnetic resonance (MR) imaging and/or contrast-enhanced computed tomography. All of the patients’ initial craniotomies for tumor resection were performed at MD Anderson. Histological diagnosis in each case was based on the World Health Organization Classification and determined by a board-certified pathologist. The patients’ demographic, clinical, and radiographic data were reviewed. Clinical variables included age at surgery, sex, and Karnofsky Performance Scale (KPS) score. Volumetric analysis was performed on immediate preoperative MR imaging. Preoperative and postoperative tumor volumes measured using MR imaging were quantified prospectively using Vitrea (version 2) or MedVision (version 1.41) software. These software programs allow for calculation of the tumor area as outlined on selected axial or coronal images followed by estimation of the tumor volume based on the known thickness of the slice. Tumor contrast-enhanced volume was defined as the area of increased signal intensity on contrast-enhancing T1-weighted MR images. Extent of resection (EOR) was calculated based on the preoperative and postoperative tumor volumes. A subset of patients had initial diagnostic biopsies at outside institutions followed by definitive resection at MD Anderson. All “second-look” surgeries were performed within 6 weeks of biopsy confirming the tumor to be GBM.
Immunohistochemical Analysis
Immunohistochemical analysis was performed using entire sections of formalin-fixed, paraffin-embedded GBM samples with automated staining techniques as we described previously.9 Dilutions and conditions were performed according to package insert instructions; they were optimized and validated and met the standards and requirements of the Clinical Laboratory Improvement Amendments, College of American Pathologists, and International Organization for Standardization. Immunohistochemistry results were evaluated independently by 6 board-certified neuropathologists. The primary anti-programmed death-ligand 1 (PD-L1) antibody SP142 (Spring Bioscience) was used for detection. The chromogenic reporter 3, 3′-diaminobenzidine was used to facilitate colorimetric visualization of the antibody, yielding a brown stain that could be analyzed under a light microscope. For an SP142 clone, the Rabbit LINKER visualization system (Dako) was used. The staining was regarded as positive if its intensity on the membranes of tumor cells was greater than 1+ on a semiquantitative scale of 0–3+ (0 for no staining, 1+ for weak cytoplasmic staining, 2+ for moderate membranous staining, and 3+ for strong membranous staining) and the percentage of positively stained cells was greater than 5%. During the validation process for each analysis using immunohistochemistry, any interpathologist variability in evaluation was addressed in a microscopic examination session by all 6 pathologists led by the medical director.
Next-Generation Sequencing Methods
Next-generation sequencing was performed with genomic DNA isolated from formalin-fixed, paraffin-embedded tumor samples on the Illumina NextSeq platform. A custom-designed SureSelect XT assay (Agilent Technologies) was used to enrich 592 whole-gene targets (http://www.carislifesciences.com). Genetic variants were detected with greater than 99% confidence interval based on allele frequency and amplicon coverage, with an average sequencing depth of coverage greater than 500× and analytic sensitivity of 5% variant frequency. Variants were classified according to the American College of Medical Genetics and Genomics guidelines.10 Pathogenic variants, defined as known pathogenic mutations that promote tumorigenesis, presumed pathogenic variants (not germline with evidence of pathogenicity in functional assays), or variants that were not wild type but could be associated with protein changes, were selected for analysis of their association with survival. Wild type and benign variants that occur during somatic development and are not capable of causing disease were not included. Tumor mutational burden (TMB) was calculated by counting all nonsynonymous missense mutations not previously reported as germline alterations. Specifically, TMB was calculated using somatic nonsynonymous missense mutations in accordance with the TMB Harmonization Project (Friends of Cancer Research; http://www.focr.org/tmb), adding nonsynonymous, nonsense, in-frame indel, and frameshift variants after filtering out presumed germline variants determined from the Genome Aggregation Database (release 2.1; https://gnomad.broadinstitute.org/), the Database of Single Nucleotide Polymorphisms (human build 151; https://www.ncbi.nlm.nih.gov/snp/), and the Caris Life Sciences in-house benign database. About 1.4 Mb per tumor sample was sequenced.11
Statistical Methods
Univariate analysis to assess the associations of individual features with STS, MTS, and LTS was performed using an ordinal proportional odds regression model. Confidence intervals (CIs) were computed using Wald statistics and P values were determined using likelihood ratio tests. Features with P values of up to .10 in the univariate analysis were subjected to multivariate analysis. To select the best features for the model, the forward stepwise selection was used with the Akaike information criterion (AIC). AIC balances how well a model explains the data (quality of predictions) with the model’s complexity (number of features). The goal is to have a model that explains data well but is not too complex. Lower AIC values are better. Forward stepwise selection starts by constructing p single-feature models in which p is the number of features under consideration. The best-fitting single-feature model, as measured by AIC, is chosen for step 2. In step 2, p−1 two-feature models are built. Each model uses the single best feature from the first step of the algorithm and one of the remaining p−1 features. The 2 features used in the best model are then selected for step 3. This process continues until all features are used or the AIC begins to increase (suggesting an excessively complex model). The results of the forward stepwise selection are shown in Supplementary Figure 1.
To address the differing prevalence of STS, MTS, and LTS in the MD Anderson cohort and GBM patient data in The Cancer Genome Atlas (TCGA), Bayes’ theorem was used. Let and be the known class prevalence for the MD Anderson and TCGA patients, respectively. Also, let be the probability of observing features given class , where class c is , , or . Then the model outputs
For making predictions on the TCGA cohort the probabilities
were performed. Our strategy was to use the known and to determine from using the first equation. was then put into the second equation (along with , which is known) to determine . Of note, can only be determined up to a multiplicative constant.
At fixed , we seek sought the vector , which minimizes
We found at each vector of probabilities in the grid using a Newton-type algorithm.
Statistical analyses were performed using R statistical software (version 3.5.1) with packages survival v3.1–8, MASS 7.3–51.1, and plotted using gplot v2_3.2.1.
Results
Characteristics of the Patient Cohort
The median age of the MD Anderson GBM patient cohort was 57 years (range, 25–81 years), their median preoperative KPS score was 90 (range, 40–100), and 63% (n = 50) of them were male (Table 1). Preoperative imaging revealed that tumor necrosis predominated in the analyzed cohort, which is consistent with the radiographic features of GBM. We performed volumetric analysis on the T1 contrast-enhancing portion of the tumor, its T2/fluid-attenuated inversion recovery (FLAIR) volume and its MRI tumor necrosis characteristics. No volumetric data were available for one patient. The median preoperative T1 contrast-enhancing disease volume was 32.1 cm3 (range, 0.4–111.0 cm3), and the median T2/FLAIR volume was 79.5 cm3 (range, 4.9–300.8 cm3). Five tumors did not show sufficient enhancement on T1-weighted sequences to be volumetrically measured, and 4 had no measurable T2 volumes. For all other patients (n = 71), we calculated the ratio of contrast-enhancing to nonenhancing disease (T1/T2 ratio), finding a median value of 41%. Tumor necrosis was present in 55 cases (69%), with a median volume of 10.6 cm3 (range, 0.2–75.5 cm3). Three patients underwent biopsy only, with the remaining 77 undergoing resection. Of these patients, 74% (n = 57) has a gross total resection (≥97% resection of T1 contrast-enhancing disease), and 66% (n = 51) had 100% resection of T1 contrast-enhancing disease. Three patients had diagnostic biopsies at outside institutions and subsequently presented to MD Anderson and underwent resection; we included these patients in the survival analysis. The overall median EOR was 100.0% (range, 50.5–100.0%) for T1 contrast-enhancing disease and 63.1% (range, 14.4–100.0%) for T2/FLAIR disease. Postoperatively, T2/FLAIR disease in one patient could not be assessed due to a large area of postoperative ischemic change that was indistinguishable from residual T2/FLAIR disease.
Table 1.
Clinical Characteristics of the GBM Patient Cohort (N = 80)
| Characteristic | n | % |
|---|---|---|
| Sex | ||
| Female | 30 | 38 |
| Male | 50 | 62 |
| Clinical trial enrollment | ||
| No | 54 | 68 |
| Yes | 25 | 31 |
| Unknown | 1 | 1 |
| Tumor necrosis (n = 77) | ||
| No | 22 | 29 |
| Yes | 55 | 71 |
| Surgical approach | ||
| Craniotomy/resection | 77 | 96 |
| Biopsy | 3 | 4 |
| EOR (n = 77) | ||
| GTR | 57 | 74 |
| STR | 20 | 26 |
EOR, extent of resection; GTR, gross total resection; STR, subtotal resection.
Clinical Factors Associated With Survival of GBM Patients
The median overall survival time in the cohort was 1.2 years, with 37 (46%) experiencing STS, 22 (28%) experiencing MTS, and 21 (26%) experiencing LTS. Consistent with reports in the literature,5,6 univariate analysis demonstrated that age was a significant predictor of overall survival (P < .0001; odds ratio [OR], 0.88 [95% CI, 0.84–0.92]; Table 2). Overall survival durations were shorter in male than in female patients (P = .0164; OR, 0.34 [95% CI, 0.14–0.85]). Additionally, patients with higher preoperative KPS scores had longer survival than did patients with lower scores (P < .0001; OR, 1.09 [95% CI, 1.04–1.14]). Clinical trial enrollment at any point in the patients’ treatment course impacted the outcome, with trial participants having a longer survival (P < .0001; OR, 6.52 [95% CI, 2.51–16.92]). Multivariant analysis showed that higher KPS was significantly associated with clinical trial participation (P = .0143). Clinical trial participation was not significantly associated with IDH1 status (P = .3271).
Table 2.
Univariate Analysis of the Study Patients (N = 80): Clinical and Radiographic Factors
| Feature | n | OR | 95% CI | P |
|---|---|---|---|---|
| Age | — | 0.8797417 | 0.84–0.92 | <.0001 |
| Sex | ||||
| Female | 30 | — | — | .0164 |
| Male | 50 | 0.3437707 | 0.14–0.85 | |
| KPS score | — | 1.0853484 | 1.04–1.14 | <.0001 |
| T1-enhancing volume | — | 0.9839941 | 0.97–1.00 | .0497 |
| T2 volume including T1 | — | 1.0027202 | 1.00–1.01 | .4802 |
| T1/T2 volumetric ratio | — | 0.0807405 | 0.01–0.45 | .0022 |
| Necrosis volume | — | 0.9699149 | 0.93–1.01 | .0817 |
| Volumetric EOR based on enhancement | — | 1.0087027 | 0.96–1.06 | .7141 |
| Volumetric EOR based on T2/FLAIR disease | — | 0.9947252 | 0.98–1.01 | .5930 |
| EOR based on enhancing disease | ||||
| GTR | 57 | — | — | .3419 |
| STR | 16 | 0.4584197 | 0.15–1.40 | |
| Biopsy | 3 | 0.5608276 | 0.04–7.37 | |
| PD-L1 status | ||||
| Negative | 46 | — | — | .7869 |
| Positive | 6 | 0.7866907 | 0.13–4.75 | |
| TMB | — | 0.7378053 | 0.58–0.94 | .0055 |
| Clinical trial enrollment | — | |||
| No | 54 | — | — | <.0001 |
| Yes | 25 | 6.5197993 | 2.51–16.92 | |
| MGMT methylation | ||||
| No | 38 | — | — | .0032 |
| Yes | 32 | 3.9444285 | 1.52–10.22 |
KPS, Karnofsky Performance Scale; EOR, extent of resection; GTR, gross total resection; STR, subtotal resection; TMB, tumor mutational burden.
The T1/T2 Ratio Impacts GBM Patient Survival
From a radiographic perspective, a higher preoperative T1 contrast-enhancing tumor volume (P = .0497; OR, 0.98 [95% CI, 0.97–1.00]) was associated with a shorter survival time. More specifically, the T1/T2 ratio correlated negatively with survival. Patients with larger T1/T2 ratios (sample patient—Figure 1) had a significantly shorter survival (P = .0022; OR, 0.08 [95% CI, 0.01–0.45]). Among the patients who experienced LTS (n = 21), volumetric data were available for 17, and of those, 15 (88%) had 100% resection of the T1-enhancing component of the tumor (median, 100.0% [range, 57.9–100.0%]). Volumetric data were available for all short-term survivors (n = 37), with 21 (57%) undergoing 100% resection (24 patients had ≥97% resection).
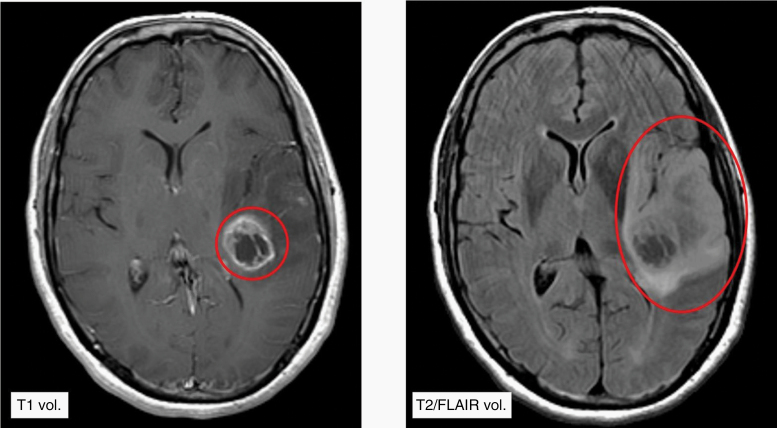
Figure 1.
Representative MR images of a newly diagnosed GBM patient at presentation demonstrating a T1 gadolinium contrast-enhancing volume of 14.2 cm3 (left) and T2/FLAIR volume of 104.4 cm3 (right). In this patient, the T1/T2 ratio was 0.13.
Genetic Determinants of GBM Patient Survival
Univariate analysis revealed that 2 gene mutations were significantly associated with survival (Table 3). IDH1 mutations had the biggest impact on survival, with patients having IDH1-mutant tumors (n = 14) surviving longer than did those having IDH1–wild-type tumors (P < .0001; OR, 97.8 [95% CI, 10.79–886.91]). Most of the long-term survivors (13 of 21) had this mutation, whereas none of the short-term survivors had it. PTEN status also influenced survival, with patients having tumors with mutations of the PTEN genes experiencing shorter survival than did those with PTEN–wild-type tumors (P = .0235; OR, 0.23 [95% CI, 0.06–0.94]). None of the IDH1-mutated patients (n = 14) had PTEN mutations (n = 11). However, the correlation was not statistically significant. PD-L1 expression was not associated with survival in our cohort.
Table 3.
Univariate Analysis of the Study Patients (N = 80): Molecular Variables
| Variable {“Gene?”} | Mutationa | n | OR | 95% CI | P |
|---|---|---|---|---|---|
| APC | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| BRAF | 0 | 78 | — | — | .6671 |
| 1 | 2 | 1.5837893 | 0.19–13.36 | ||
| CDKN2A | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| EGFR | 0 | 67 | — | — | .3197 |
| 1 | 13 | 0.5516976 | 0.16–1.86 | ||
| FBXW7 | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| FGFR3 | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| IDH1 | 0 | 66 | — | — | <.0001 |
| 1 | 14 | 97.8096211 | 10.79–886.91 | ||
| KRAS | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| MUTYH | 0 | 78 | — | — | .6769 |
| 1 | 2 | 0.5930654 | 0.05–7.64 | ||
| NF1 | 0 | 75 | — | — | .3161 |
| 1 | 5 | 0.4264895 | 0.07–2.50 | ||
| PIK3CA | 0 | 73 | — | — | .4826 |
| 1 | 7 | 1.6332647 | 0.41–6.59 | ||
| PTEN | 0 | 69 | — | — | .0235 |
| 1 | 11 | 0.2313771 | 0.06–0.94 | ||
| PTPN11 | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| SUFU | 0 | 79 | — | — | |
| 1 | 1 | — | — | ||
| TP53 | 0 | 57 | — | — | .0622 |
| 1 | 23 | 2.4522865 | 0.93–6.45 |
a0, no mutation; 1, mutation.
Consolidating the overall mutational landscape, we evaluated the association of TMB with survival and found that a high TMB negatively impacted survival (P = .0055; OR, 0.73 [95% CI, 0.58–0.94]). IDH1 mutation was significantly associated with lower TMB (P = .0003). Furthermore, O[6]-methylguanine-DNA methyltransferase (MGMT) methylation was associated with LTS (P = .0032; OR, 3.94 [95% CI, 1.52–10.22]).
Construction of the Predictive Nomogram
The dataset was missing the following variables for some patients: PD-L1 expression (n = 28), MGMT methylation status (n = 10), T1/T2 ratio (n = 9), volumetric EOR of enhancing disease (n = 7), volumetric EOR of T2/FLAIR disease (n = 7), TMB (n = 6), T1-enhancing volume (n = 5), EOR of enhancing disease (n = 4), T2 volume including T1 (n = 4), necrosis volume (n = 3), KPS score (n = 2), and clinical trial participation (n = 1). For PD-L1 tumor staining and TMB analysis, these assays were becoming available as a Clinical Laboratory Improvement Amendments approved assay during data acquisition. The remaining missing variables were mostly related to the inability to precisely measure multifocal and complex images. Genomic variables were coded as unfavorable if they were considered pathogenic or presumed pathogenic based on the American College of Medical Genetics and Genomics guidelines.10 We coded all other genetic alterations, such as benign, variant not detected, and variant of unknown significance, as neutral. Thus all genomic variables were binary. We then correlated each variable with STS, MTS, and LTS and ranked based on the significance of each correlation. We considered all variables with P values less than .1 in the univariate analysis to be potentially interesting.
We found that clinical trial enrollment correlated with survival. However, we eliminated this feature from the survival-predictive nomogram because clinical trial participation cannot be determined at the time of GBM diagnosis. We also removed TMB from our survival-predictive nomogram because its definition varies from assay to assay, making reproduction with an independent dataset such as those in the TCGA difficult.
Model Building
We used a proportional odds ordinal logistic regression model to predict STS, MTS, and LTS probability given according to patient features (covariates).12 We used forward stepwise selection with the AIC to select the best features for the model implemented using the MASS package in R statistical software.13 We used age, IDH1, KPS score, PTEN, and TP53 mutation status in the final model. The genomic variables IDH1, PTEN, and TP53 are all binary (either mutated or not mutated). Supplementary Figure 1 shows that the model assigned low probability of LTS to the patients who actually experienced STS (concentration of red region near 0) and high probability of LTS to patients who actually experienced LTS (blue region mode near 1). The code used for building the model can be found at https://github.com/longjp/GBMpredict. Statistical details of the proportional odds model, AIC, and forward stepwise selection are described above.
Evaluation and Validation of the Survival-Predictive Nomogram
We externally validated the model using GBM patient data from TCGA (n = 592) with the following patient/survival proportions: STS (0–1 year), 0.42; MTS (1–5 years), 0.52; and LTS (5+ years), 0.06. These proportions differed markedly from the proportions in our cohort, in which patients with LTS were deliberately oversampled to obtain enough patients to build a robust model (STS, 0.46; MTS, 0.28; LTS, 0.26). To account for these differences in survival prevalence, we adjusted our probabilistic predictions to reflect the prevalence in TCGA using Bayes’ theorem as described above.
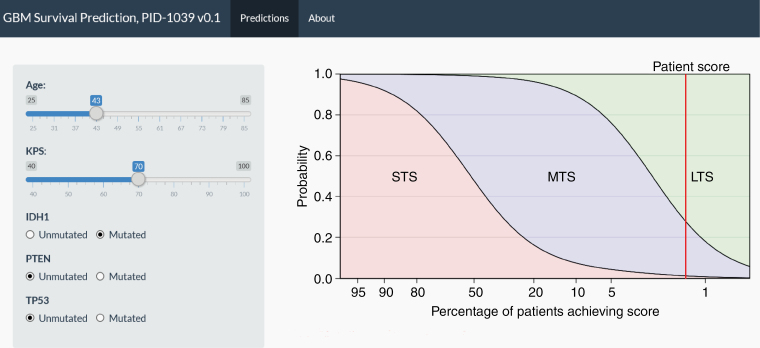
We then applied the model to the TCGA cohort and made probabilistic predictions for each patient. For any given value of age, KPS score, IDH1, PTEN, and TP53 mutation status, we obtained a set of 3 probabilities corresponding to the likelihood of STS, MTS, and LTS. Since 95 patients in the TCGA cohort are censored (patients are STS or MTS but alive at the time of the last follow-up so true survival is at least STS or MTS), we use the concordance index to assess the quality of these predictions.12 The C-index is the proportion of comparable pairs of samples where the model predictions agree with the actual outcomes. A C-index of 0.5 corresponds to a useless model (random guessing) and 1.0 to a perfect model. Our model achieves a C-index of 0.66 on the independent TCGA cohort (P < .00001). The finalized embargoed nomogram can be found at https://biostatistics.mdanderson.org/shinyapps/GBM_Predict/ (username: GBM; password: predict; Figure 2).
Figure 2.
The application for GBM survival prediction using the survival-predictive nomogram. A representative clinical example is shown in which a 43-year-old GBM patient with a KPS score of 70 had an IDH1-mutant tumor. This patient had a 2%, 72%, and 26% chance of being a short-term survivor (<6 months), long-term survivor (>5 years), and median-term survivor (~15 months), respectively. Two percent of patients had this outcome.
Discussion
Long-term survival in patients with GBM is uncommon, and survival for at least 5 years is quite rare. In this report, we describe clinical, radiographic, and molecular features associated with LTS in this patient population. These key features should be accounted for during clinical trial interpretation as confounders when assessing outcomes in GBM patients. Notably, 50% of the long-term survivors in our study participated in at least one clinical trial. This may explain why the initial results of many uncontrolled single-institution GBM studies appear to be promising but the effect is diminished once the studies are expanded to multi-institutional controlled trials. Researchers have shown that clinical features such as young age and a good KPS score at the time of diagnosis are favorable prognostic factors in GBM patients.14–16 Because long-term survivors represent only a small subset of GBM patients, their clinical characteristics were only minimally addressed in earlier studies, often with limited sample sizes and follow-up durations.17–21 In one study, investigators evaluated 55 patients with GBM who underwent surgery and adjuvant radiotherapy and survived beyond 36 months.6 That study demonstrated that LTS was associated with high functional status and young age. Another retrospective analysis compared 16 long-term survivors who survived for more than 2 years with short- and medium-term survivors and found that KPS score at diagnosis, age, initial tumor resection (as opposed to biopsy), chemoradiation, and early progression were factors associated with survival in these groups.17 However, few studies22 account for radiographic, clinical, and molecular determinants of the outcome as in the present study. Although researchers have attempted to correlate features of advanced imaging modalities such as radiomic and textural MR imaging features with GBM outcome,23–25 the present study shows the utility of conventional standard MR imaging in assessing the outcome of GBM. An advantage of the nomogram is the ease of interrogation of outcome in the clinical domain relative to more complex mathematical modeling even if the latter is more accurate.26 Furthermore, our analysis demonstrated that the influence of a pathogenic mutation of IDH1, which is directly associated with age, surpassed that of many other previously documented factors. One could argue that the presence of an IDH1 mutation may demonstrate that these cases are not classical GBM. There is a growing consensus that IDH-mutant astrocytomas and IDH-wildtype, grade 4 diffuse astrocytomas are 2 biologically distinct tumors.27,28 Notably, board-certified neuropathologists who advise regarding the World Health Organization criterion reviewed all of the cases, none of the analyzed cases were secondary GBM, and IDH1 mutations were present in about 5% of cases in a previous study.29
Because the median EOR in our study cohort was 100%, an excellent resection is a baseline criterion for subsequent application of the survival-predictive nomogram. In a recently published GBM nomogram, in addition to age, KPS, and gender, unlike the current study, these authors included EOR to their internally validated nomogram.30 Gorlia et al.31 published a large study compiling data from 573 GBM patients to construct a predictive nomogram for 2-year survival with a focus on the impact of MGMT status. However, at that time no validation set was available and our current study has a greater depth of molecular profiling. Genetic analysis provided the compelling perspective that the presence of a pathogenic IDH1 mutation is one of the strongest influences on LTS in GBM patients. To account for the possibility of inclusion of patients with secondary GBM in our LTS cohort, we performed genetic analysis of the long-term survivors that demonstrated all of their tumors had intact ATRX and that none of them had previously documented gliomas, indicating that these were de novo GBMs. None of the IDH-mutant tumors showed the classic molecular alteration triad of IDH mutation, TP53 mutation, and ATRX mutation, which represents a rare subtype. Among prior profiling of IDH mutants that are non-co-deleted, approximately 95% are TP53 and 75% are ATRX mutant.32,33 We validated the GBM survival-predictive nomogram using GBM patient data in TCGA because validation in other large cases series was not possible, as outcome data were only reported for up to 3 years, the numbers of 5-year survivors lacked sufficient statistical power, and/or genomic data were insufficient.5,34–37 In the future, more molecular information on patient GBMs will be available both because it is a requirement for diagnosis using World Health Organization criteria and because genetic profiling is becoming more mainstream. Therefore, we will update the nomogram in the future to enhance its accuracy in survival prediction.
There are some study limitations that warrant discussion. The nomogram was developed based on the data from an academic institution, and although validated with TCGA, may not be reflective of patients treated in a general community practice setting. Biases such as the overall surgical EOR from the academic group was notably high and may not be recapitulated without neuro-oncology neurosurgical expertise. Despite the extensive tumor profiling performed, the patient cohort itself is not large relative to more common cancers. Additionally, epigenetic profiling would further enhance the predictive value of the nomogram but this is not routinely performed on clinical specimens at this time.
In summary, this study demonstrated a validated integration of clinical, radiographic, and genomic data to predict the survival of GBM. Note, our C-index of 0.66 compares favorably with 2 other published GBM nomograms (0.65730 and 0.69538). A C-index of 0.66 indicates additional opportunities to improve the predictive capabilities of this nomogram likely based on epigenetic profiling, but they have yet to be used for clinical profiling in GBM patients. Although prognoses for GBM remain poor overall, this study does provide optimism for a subgroup of patients. Our new LTS-predictive score can provide clinicians with an easier way to inform patients about their relative chances of LTS. These findings may also assist clinical trial investigators as well as regulatory agencies and other stakeholders in identifying confounding variables influencing the interpretation of GBM outcome data in nonrandomized clinical trials.
Supplementary Material
Acknowledgments
Special thanks to David M. Wildrick, PhD, Donald R. Norwood, and Audria Patrick for their editorial and administrative support.
Funding
This study was supported by the National Institutes of Health/National Cancer Institute under award numbers CA120813 and P30CA016672, the Provost Retention Fund, and Golfers Against Cancer.
Conflict of interest statement. S.D.F., T.H., N.K.M., K.A.M., W.N.A., D.S., J.F.G., G.N.F., G.R., R.V., R.S., and J.P.L. have no disclosures to report. A.B.H. serves on the advisory boards of Caris Life Sciences and WCG Oncology, receives royalties on licensed intellectual property from Celldex Therapeutics and DNAtrix, and receives research support from Celularity, Carthera, Codiak, and Moleculin. J.X. and D.S. are employed by Caris Life Sciences and D.S. holds stock in Caris Life Sciences. M.K. is a consultant for AbbVie, Ipsen, Pfizer Roche, and Jackson Laboratory for Genomic Medicine; research funding from AbbVie, Bristol-Myers Squibb (BMS), and Specialized Therapeutics.
Authorship Statement:
Conception or design of the work: S.D.F., T.H., J.P.L., and A.B.H.; data collection: S.D.F., T.H., L.X., M.L., R.C., C.J., and A.B.H.; data analysis and interpretation: S.D.F., T.H., G.N.F., J.P.L., and A.B.H.; drafting the article: S.D.F., T.H., J.F.G., J.T., G.R., D.S., M.K., J.P.L., and A.B.H.; critical revision of the article: S.D.F., T.H., N.K.M., K.A.M., W.N.A., D.S., J.F.G., G.N.F., G.R., J.X., R.V., D.S., M.K., R.S., J.P.L., and A.B.H.; final approval of the version to be published: S.D.F., T.H., N.K.M., K.A.M., W.N.A., D.S., J.F.G., G.N.F., L.X., M.L., G.R., R.C., J.X., R.V., D.S., M.K., R.S., J.P.L., and A.B.H.
Data Availability
De-identified individual participant data can be made available after publication by request to the corresponding author. A research proposal should be included.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups ; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chandler KL, Prados MD, Malec M, Wilson CB. Long-term survival in patients with glioblastoma multiforme. Neurosurgery. 1993;32(5):716–720; discussion 720. [DOI] [PubMed] [Google Scholar]
- 5. Krex D, Klink B, Hartmann C, et al. ; German Glioma Network Long-term survival with glioblastoma multiforme. Brain. 2007;130(Pt 10):2596–2606. [DOI] [PubMed] [Google Scholar]
- 6. Cantero D, Rodríguez de Lope Á, Moreno de la Presa R, et al. Molecular study of long-term survivors of glioblastoma by gene-targeted next-generation sequencing. J Neuropathol Exp Neurol. 2018;77(8):710–716. [DOI] [PubMed] [Google Scholar]
- 7. Lu J, Cowperthwaite MC, Burnett MG, Shpak M. Molecular predictors of long-term survival in glioblastoma multiforme patients. PLoS One. 2016;11(4):e0154313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng S, Dhruv H, Armstrong B, et al. Integrated genomic analysis of survival outliers in glioblastoma. Neuro Oncol. 2017;19(6):833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garber ST, Hashimoto Y, Weathers SP, et al. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro Oncol. 2016;18(10):1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agresti A. Categorical Data Analysis. Hoboken, NJ: John Wiley & Sons, Inc; 2003. [Google Scholar]
- 13. Venables WN, Ripley BD.. Modern Applied Statistics With S-PLUS. New York, NY: Springer Science & Business Media; 2013. [Google Scholar]
- 14. Wick W, Platten M, Meisner C, et al. ; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 15. Barker FG 2nd, Chang SM, Larson DA, et al. Age and radiation response in glioblastoma multiforme. Neurosurgery. 2001;49(6):1288–1297; discussion 1297. [DOI] [PubMed] [Google Scholar]
- 16. Gittleman H, Lim D, Kattan MW, et al. An independently validated nomogram for individualized estimation of survival among patients with newly diagnosed glioblastoma: NRG Oncology RTOG 0525 and 0825. Neuro Oncol. 2017;19(5):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mazaris P, Hong X, Altshuler D, et al. Key determinants of short-term and long-term glioblastoma survival: a 14-year retrospective study of patients from the Hermelin Brain Tumor Center at Henry Ford Hospital. Clin Neurol Neurosurg. 2014;120:103–112. [DOI] [PubMed] [Google Scholar]
- 18. Chaichana K, Parker S, Olivi A, Quiñones-Hinojosa A. A proposed classification system that projects outcomes based on preoperative variables for adult patients with glioblastoma multiforme. J Neurosurg. 2010;112(5):997–1004. [DOI] [PubMed] [Google Scholar]
- 19. Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro Oncol. 2004;6(3):227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marko NF, Toms SA, Barnett GH, Weil R. Genomic expression patterns distinguish long-term from short-term glioblastoma survivors: a preliminary feasibility study. Genomics. 2008;91(5):395–406. [DOI] [PubMed] [Google Scholar]
- 21. Burton EC, Lamborn KR, Feuerstein BG, et al. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res. 2002;62(21):6205–6210. [PubMed] [Google Scholar]
- 22. Molinaro AM, Hervey-Jumper S, Morshed RA, et al. Association of maximal extent of resection of contrast-enhanced and non-contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol. 2020;6(4):495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bae S, Choi YS, Ahn SS, et al. Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology. 2018;289(3):797–806. [DOI] [PubMed] [Google Scholar]
- 24. Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280(3):880–889. [DOI] [PubMed] [Google Scholar]
- 25. Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys. 2015;42(11):6725–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ponnapalli SP, Bradley MW, Devine K, et al. Retrospective clinical trial experimentally validates glioblastoma genome-wide pattern of DNA copy-number alterations predictor of survival. APL Bioeng. 2020;4(2):026106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Louis DN, Wesseling P, Aldape K, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30(4):844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu A, Hou C, Chen H, Zong X, Zong P. Genetics and epigenetics of glioblastoma: applications and overall incidence of IDH1 mutation. Front Oncol. 2016;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gittleman H, Cioffi G, Chunduru P, et al. An independently validated nomogram for isocitrate dehydrogenase-wild-type glioblastoma patient survival. Neurooncol Adv. 2019;1(1):vdz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol. 2008;9(1):29–38. [DOI] [PubMed] [Google Scholar]
- 32. Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse Glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hottinger AF, Yoon H, DeAngelis LM, Abrey LE. Neurological outcome of long-term glioblastoma survivors. J Neurooncol. 2009;95(3):301–305. [DOI] [PubMed] [Google Scholar]
- 35. McLendon RE, Halperin EC. Is the long-term survival of patients with intracranial glioblastoma multiforme overstated? Cancer. 2003;98(8):1745–1748. [DOI] [PubMed] [Google Scholar]
- 36. Polley MY, Lamborn KR, Chang SM, Butowski N, Clarke JL, Prados M. Conditional probability of survival in patients with newly diagnosed glioblastoma. J Clin Oncol. 2011;29(31):4175–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups ; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 38. Molitoris JK, Rao YJ, Patel RA, et al. Multi-institutional external validation of a novel glioblastoma prognostic nomogram incorporating MGMT methylation. J Neurooncol. 2017;134(2):331–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified individual participant data can be made available after publication by request to the corresponding author. A research proposal should be included.