Figure 1.
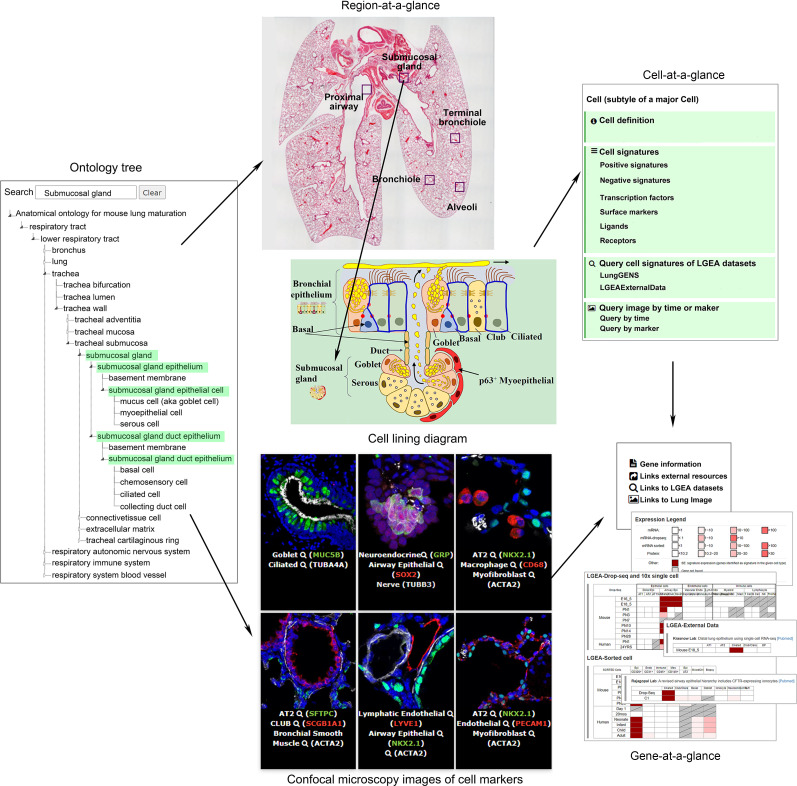
“Lung-at-a-glance” consists of the following three interactive and interconnected components: “region-at-a-glance,” “cell-at-a-glance,” and “gene-at-a-glance.” Region-at-a-glance enables users to search a specific lung region using the interactive ontology tree navigation or by clicking the boxes in the lung image to explore cells residing in the selected region. Cell-at-a-glance offers a collection of information on queried cell types, including cell definition, cell type–specific positive and negative markers, transcription factors, ligand receptors, and hyperlinks that are mapped to the chosen cell type in all datasets in the Lung Gene Expression Analysis (LGEA) web portal and immunofluorescence confocal images of the chosen cell type. Gene-at-a-glance allows users to query a gene of interest to obtain associated gene information, including hyperlinks to external knowledge bases, expression patterns in LGEA datasets, and immunofluorescence confocal images associated with the gene marker. A two-dimensional heatmap organized by developmental times and cell types was used to summarize expression patterns and statistics of queried genes in all datasets available in LGEA databases. Screen images are taken from the LGEA web portal (https://research.cchmc.org/pbge/lunggens/mainportal.html). Adapted from Reference 16. LYVE1 = lymphatic vessel endothelial hyaluronan receptor 1; NKX2.1 = NK2 homeobox 1; PECAM1 = platelet and endothelial cell adhesion molecule 1; SOX2 = SRY-box transcription factor; TUBA4A = tubulin alpha 4a; TUBB3 = tubulin beta 3 class III.