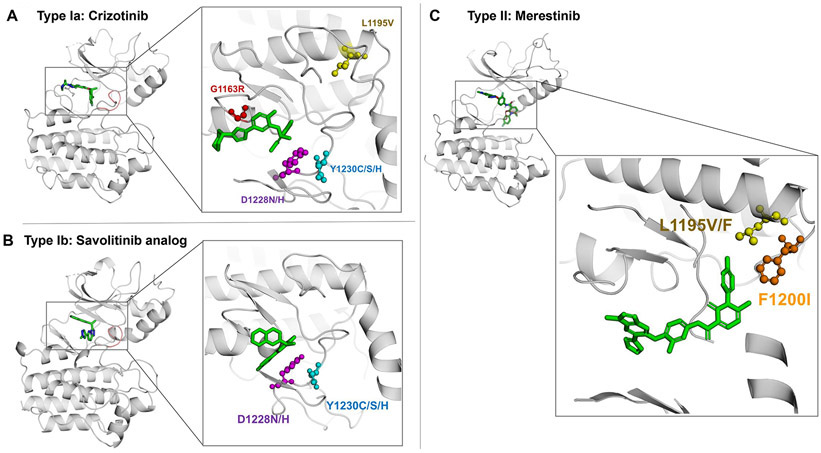
Figure 2. Mechanisms of resistance to MET tyrosine kinase inhibitors in MET exon 14 mutant lung cancer.
Crystal modelling of the MET kinase domain and binding of MET tyrosine kinase inhibitors (green). The figure displays the position of frequently mutated residues within the MET kinase domain including G1163 (red), D1228 (purple), Y1230 (blue), L1195 (yellow), and F1200 (orange) that confer acquired resistance to MET TKIs. Panel A displays interaction of the type Ia MET TKI crizotinib (PDB Ref: 2WGJ) with commonly mutated residues that confer resistance to crizotinib like G1163R, D1228X and Y1230X mutations. Panel B shows the interaction of the type Ib MET inhibitor savolitinib analog (PDB Ref: 3ZC5) with resistance mutations including D1228X and Y1230X, but the interaction with the kinase domain is not predicted to be affected by the G1163R solvent front mutation. In panel C, the type II MET TKI merestinib (PDB Ref: 4EEV) is simulated bound to the kinase domain and displays the interaction of type II MET inhibitors with key residues that can cause resistance to these compounds like L1195F/V and F1200L.