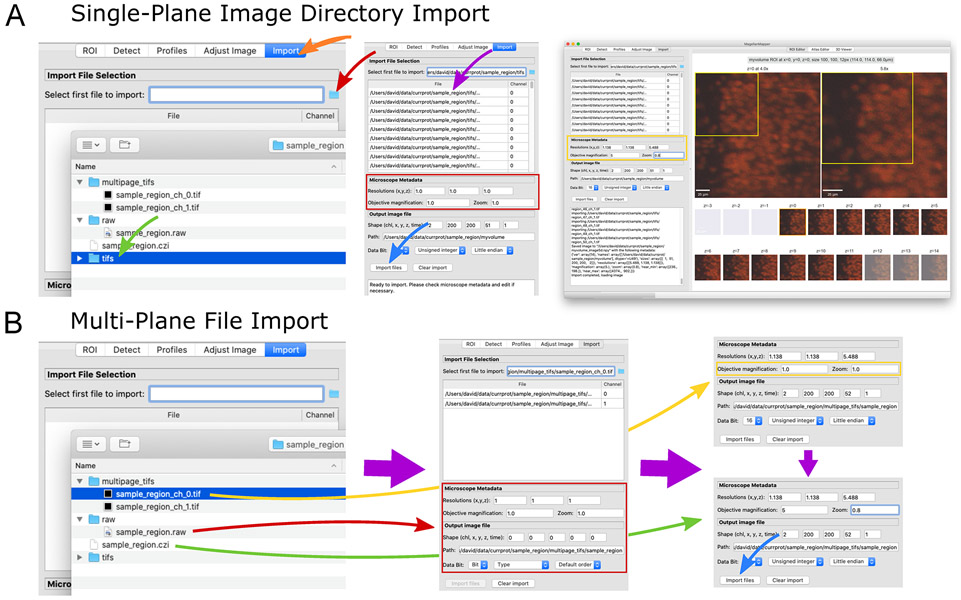
Figure 4.
Import image files into a Numpy volumetric format. (A) The “Import” panel (left, orange arrow) displays selectors for choosing a file (red arrow), such as a directory of images (green arrow). The files are loaded into the table (middle, purple arrow) with channel determined based on the filename. Microscopy metadata can be entered (red box) and pressing the “Import files” button loads (blue arrow) each image file into a separate plane of a single volumetric image file (right). (B) Several types of multi-plane images can be loaded (left). Raw image formats typically contain no metadata (red arrow), and all metadata should be entered manually. For multi-plane, multichannel TIFF files, selecting the first file (yellow arrow) will cause related files to be loaded into the table (middle). In the metadata area (middle, red box), image parameters will be loaded based on available metadata embedded in the file. In this case, partial metadata is loaded, and the rest needs to be entered manually (right, yellow box). Some proprietary formats (green arrow) contain all channels and metadata in a single file, allowing immediate import (right, blue arrow).